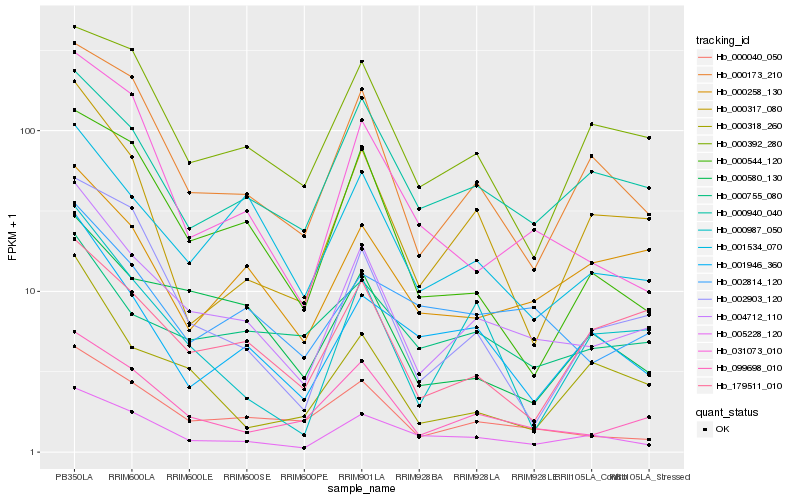
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004712_110 |
0.0 |
- |
- |
30S ribosomal protein S10, putative [Ricinus communis] |
| 2 |
Hb_099698_010 |
0.1199128836 |
- |
- |
- |
| 3 |
Hb_000040_050 |
0.1290713844 |
- |
- |
PREDICTED: uncharacterized protein LOC102617857 isoform X1 [Citrus sinensis] |
| 4 |
Hb_000173_210 |
0.142154059 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 5 |
Hb_005228_120 |
0.142489131 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 6 |
Hb_001534_070 |
0.1440528559 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 7 |
Hb_000940_040 |
0.1483652487 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 8 |
Hb_000580_130 |
0.1594669418 |
- |
- |
hypothetical protein JCGZ_14541 [Jatropha curcas] |
| 9 |
Hb_002814_120 |
0.1599368597 |
- |
- |
PREDICTED: uncharacterized protein LOC105633463 [Jatropha curcas] |
| 10 |
Hb_001946_360 |
0.1649861213 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 11 |
Hb_000317_080 |
0.1675680259 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 12 |
Hb_031073_010 |
0.1684892788 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Jatropha curcas] |
| 13 |
Hb_000392_280 |
0.1689122964 |
- |
- |
PREDICTED: pectinesterase 3 [Jatropha curcas] |
| 14 |
Hb_000258_130 |
0.1695689751 |
- |
- |
- |
| 15 |
Hb_000755_080 |
0.169981133 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 16 |
Hb_000544_120 |
0.170180319 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 17 |
Hb_000987_050 |
0.1717191317 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002903_120 |
0.1717857197 |
- |
- |
- |
| 19 |
Hb_179511_010 |
0.1730518407 |
- |
- |
- |
| 20 |
Hb_000318_260 |
0.1735872174 |
- |
- |
PREDICTED: cell division cycle-associated 7-like protein [Jatropha curcas] |