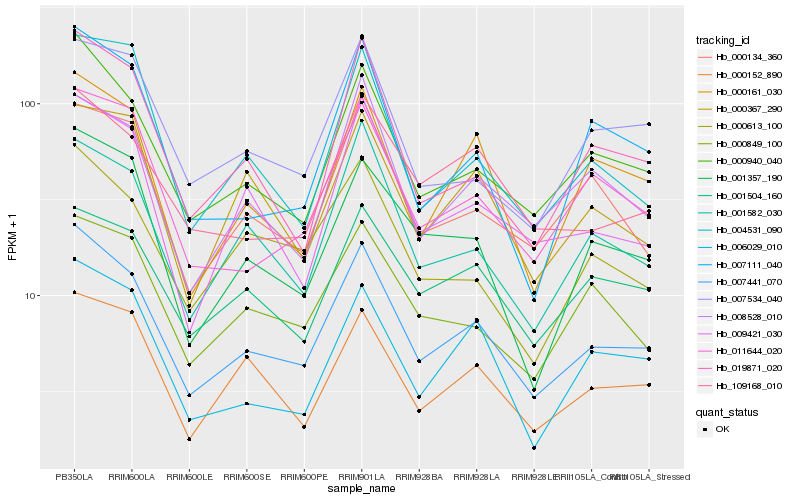
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_070 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa004494mg [Prunus persica] |
| 2 |
Hb_000940_040 |
0.0652059657 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 3 |
Hb_011644_020 |
0.0962944267 |
- |
- |
PREDICTED: choline-phosphate cytidylyltransferase 2 [Vitis vinifera] |
| 4 |
Hb_019871_020 |
0.0967222523 |
- |
- |
PREDICTED: uncharacterized protein LOC105642047 [Jatropha curcas] |
| 5 |
Hb_001582_030 |
0.096978121 |
- |
- |
PREDICTED: uncharacterized protein LOC105639462 [Jatropha curcas] |
| 6 |
Hb_001357_190 |
0.0994986398 |
- |
- |
ribonuclease III, putative [Ricinus communis] |
| 7 |
Hb_000152_890 |
0.0995272815 |
- |
- |
hypothetical protein JCGZ_26265 [Jatropha curcas] |
| 8 |
Hb_000161_030 |
0.1009153545 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP8 [Jatropha curcas] |
| 9 |
Hb_009421_030 |
0.1009394068 |
- |
- |
PREDICTED: uncharacterized protein LOC105639700 [Jatropha curcas] |
| 10 |
Hb_000134_360 |
0.1042914248 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 11 |
Hb_008528_010 |
0.1054151434 |
- |
- |
PREDICTED: uncharacterized protein LOC105628498 [Jatropha curcas] |
| 12 |
Hb_109168_010 |
0.1063273138 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 13 |
Hb_000613_100 |
0.1065673748 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 8 [Jatropha curcas] |
| 14 |
Hb_006029_010 |
0.1068222786 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 15 |
Hb_007111_040 |
0.1083562861 |
- |
- |
PREDICTED: uncharacterized protein LOC105628142 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000849_100 |
0.1088578637 |
- |
- |
PREDICTED: uncharacterized protein LOC105644134 [Jatropha curcas] |
| 17 |
Hb_004531_090 |
0.1103329459 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 18 |
Hb_001504_160 |
0.1104247239 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 19 |
Hb_007534_040 |
0.1110622011 |
- |
- |
PREDICTED: uncharacterized protein LOC105635538 [Jatropha curcas] |
| 20 |
Hb_000367_290 |
0.111582242 |
- |
- |
PREDICTED: uncharacterized protein LOC105640583 [Jatropha curcas] |