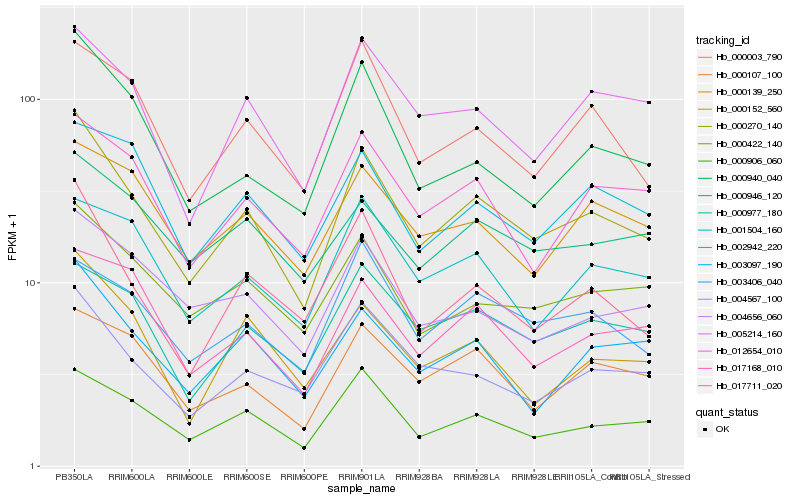
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000906_060 |
0.0955727368 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g08820 [Populus euphratica] |
| 3 |
Hb_017711_020 |
0.0998781799 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005214_160 |
0.1048896266 |
- |
- |
PREDICTED: heat shock 70 kDa protein, mitochondrial [Jatropha curcas] |
| 5 |
Hb_003097_190 |
0.1052913657 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 6 |
Hb_000422_140 |
0.1062167021 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004567_100 |
0.1066687098 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 8 |
Hb_012654_010 |
0.1076386054 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 9 |
Hb_000940_040 |
0.1091209573 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 10 |
Hb_000107_100 |
0.1102460368 |
- |
- |
hypothetical protein JCGZ_16208 [Jatropha curcas] |
| 11 |
Hb_000003_790 |
0.111512175 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 12 |
Hb_000977_180 |
0.1145916022 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000152_560 |
0.1170173367 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 14 |
Hb_017168_010 |
0.1177396051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004656_060 |
0.1185694453 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 16 |
Hb_000139_250 |
0.1203865582 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 17 |
Hb_002942_220 |
0.120531668 |
- |
- |
PREDICTED: uncharacterized protein LOC105648740 [Jatropha curcas] |
| 18 |
Hb_000946_120 |
0.1218242141 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_003406_040 |
0.122142789 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 20 |
Hb_001504_160 |
0.1230951202 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |