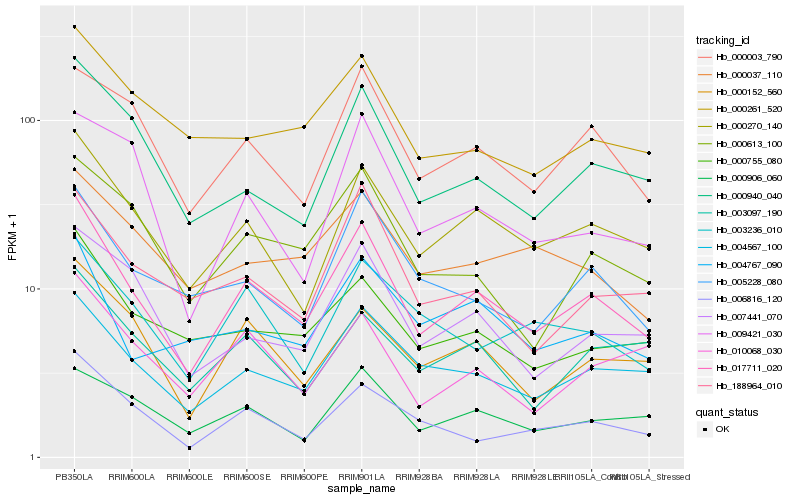
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017711_020 |
0.0 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000270_140 |
0.0998781799 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004567_100 |
0.1088010435 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 4 |
Hb_000003_790 |
0.1211472283 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 5 |
Hb_000940_040 |
0.1215333553 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 6 |
Hb_000152_560 |
0.1231120969 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 7 |
Hb_007441_070 |
0.1292232304 |
- |
- |
hypothetical protein PRUPE_ppa004494mg [Prunus persica] |
| 8 |
Hb_000755_080 |
0.1298695186 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 9 |
Hb_005228_080 |
0.1299495459 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-I [Jatropha curcas] |
| 10 |
Hb_000613_100 |
0.1311635798 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 8 [Jatropha curcas] |
| 11 |
Hb_000906_060 |
0.1335960869 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g08820 [Populus euphratica] |
| 12 |
Hb_188964_010 |
0.1336865473 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 13 |
Hb_006816_120 |
0.1341057602 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 14 |
Hb_004767_090 |
0.1372353817 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000261_520 |
0.1390123785 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 16 |
Hb_003236_010 |
0.1400523974 |
- |
- |
PREDICTED: uncharacterized protein LOC105635957 [Jatropha curcas] |
| 17 |
Hb_003097_190 |
0.1404284448 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 18 |
Hb_010068_030 |
0.1414412678 |
- |
- |
aconitase, putative [Ricinus communis] |
| 19 |
Hb_009421_030 |
0.144365701 |
- |
- |
PREDICTED: uncharacterized protein LOC105639700 [Jatropha curcas] |
| 20 |
Hb_000037_110 |
0.1450367793 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Populus euphratica] |