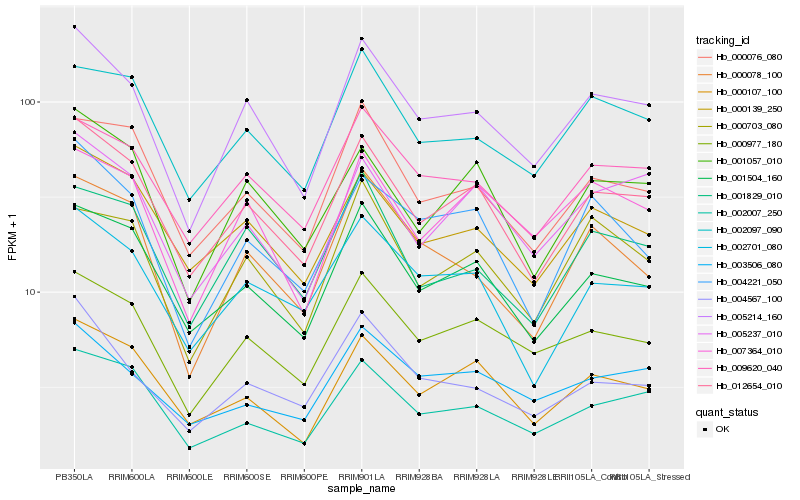
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005214_160 |
0.0 |
- |
- |
PREDICTED: heat shock 70 kDa protein, mitochondrial [Jatropha curcas] |
| 2 |
Hb_012654_010 |
0.0683574122 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 3 |
Hb_000977_180 |
0.0725276885 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_009620_040 |
0.0797639656 |
- |
- |
PREDICTED: cyclin-T1-3-like [Jatropha curcas] |
| 5 |
Hb_007364_010 |
0.0884881788 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004567_100 |
0.0887119027 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 7 |
Hb_002097_090 |
0.0890756508 |
- |
- |
Splicing factor 3A subunit, putative [Ricinus communis] |
| 8 |
Hb_001504_160 |
0.0904493901 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 9 |
Hb_000139_250 |
0.0930172865 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 10 |
Hb_002007_250 |
0.0936652132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001829_010 |
0.0943832549 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000076_080 |
0.0964005963 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 13 |
Hb_000078_100 |
0.0968353125 |
- |
- |
PREDICTED: topless-related protein 4-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000703_080 |
0.0970038081 |
- |
- |
guanine nucleotide exchange factor P532, putative [Ricinus communis] |
| 15 |
Hb_000107_100 |
0.0975586982 |
- |
- |
hypothetical protein JCGZ_16208 [Jatropha curcas] |
| 16 |
Hb_005237_010 |
0.0986858122 |
- |
- |
PREDICTED: uncharacterized protein LOC105630921 [Jatropha curcas] |
| 17 |
Hb_002701_080 |
0.0990386345 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |
| 18 |
Hb_001057_010 |
0.0993742267 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 19 |
Hb_003506_080 |
0.0994297535 |
- |
- |
dipeptidyl peptidase IV, putative [Ricinus communis] |
| 20 |
Hb_004221_050 |
0.1004322927 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |