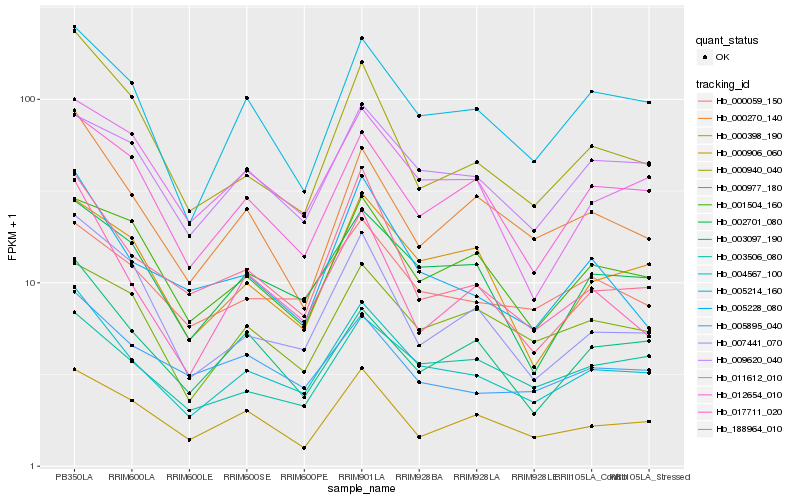
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004567_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 2 |
Hb_005214_160 |
0.0887119027 |
- |
- |
PREDICTED: heat shock 70 kDa protein, mitochondrial [Jatropha curcas] |
| 3 |
Hb_002701_080 |
0.0969553734 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |
| 4 |
Hb_012654_010 |
0.102006493 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 5 |
Hb_000059_150 |
0.1022868009 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |
| 6 |
Hb_003506_080 |
0.1054940393 |
- |
- |
dipeptidyl peptidase IV, putative [Ricinus communis] |
| 7 |
Hb_000270_140 |
0.1066687098 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 8 |
Hb_017711_020 |
0.1088010435 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005228_080 |
0.1091860593 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-I [Jatropha curcas] |
| 10 |
Hb_005895_040 |
0.1092831975 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 11 |
Hb_188964_010 |
0.1110053935 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |
| 12 |
Hb_000977_180 |
0.111321828 |
- |
- |
PREDICTED: splicing factor U2AF-associated protein 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009620_040 |
0.1114465929 |
- |
- |
PREDICTED: cyclin-T1-3-like [Jatropha curcas] |
| 14 |
Hb_000940_040 |
0.111862321 |
- |
- |
PREDICTED: protein XAP5 CIRCADIAN TIMEKEEPER isoform X1 [Jatropha curcas] |
| 15 |
Hb_000398_190 |
0.1119802395 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000906_060 |
0.1127541359 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g08820 [Populus euphratica] |
| 17 |
Hb_001504_160 |
0.1132490347 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 18 |
Hb_011612_010 |
0.1138618304 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 19 |
Hb_007441_070 |
0.11780153 |
- |
- |
hypothetical protein PRUPE_ppa004494mg [Prunus persica] |
| 20 |
Hb_003097_190 |
0.1193602825 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |