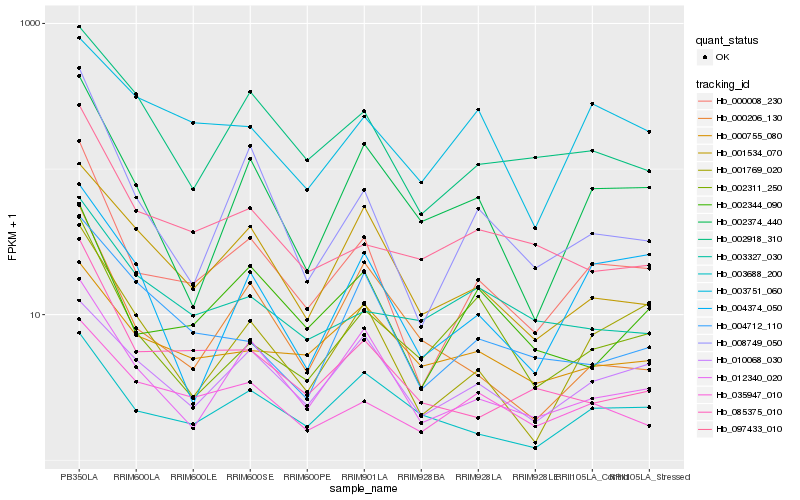
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_230 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |
| 2 |
Hb_008749_050 |
0.1400953565 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C4H3.01 [Jatropha curcas] |
| 3 |
Hb_001769_020 |
0.1529366718 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_012340_020 |
0.1532624992 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002344_090 |
0.1546496121 |
- |
- |
PREDICTED: uncharacterized protein LOC105637976 [Jatropha curcas] |
| 6 |
Hb_035947_010 |
0.165088908 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 7 |
Hb_002374_440 |
0.1659451917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_085375_010 |
0.1671927533 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_002918_310 |
0.1672322413 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 10 |
Hb_097433_010 |
0.1746501925 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TDX [Jatropha curcas] |
| 11 |
Hb_003688_200 |
0.179653119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002311_250 |
0.180171414 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 13 |
Hb_010068_030 |
0.182220177 |
- |
- |
aconitase, putative [Ricinus communis] |
| 14 |
Hb_001534_070 |
0.1864663073 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 15 |
Hb_004374_050 |
0.1915742044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004712_110 |
0.1945592452 |
- |
- |
30S ribosomal protein S10, putative [Ricinus communis] |
| 17 |
Hb_000206_130 |
0.1995339445 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_003327_030 |
0.2008071207 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |
| 19 |
Hb_000755_080 |
0.203174436 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 20 |
Hb_003751_060 |
0.2043896378 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |