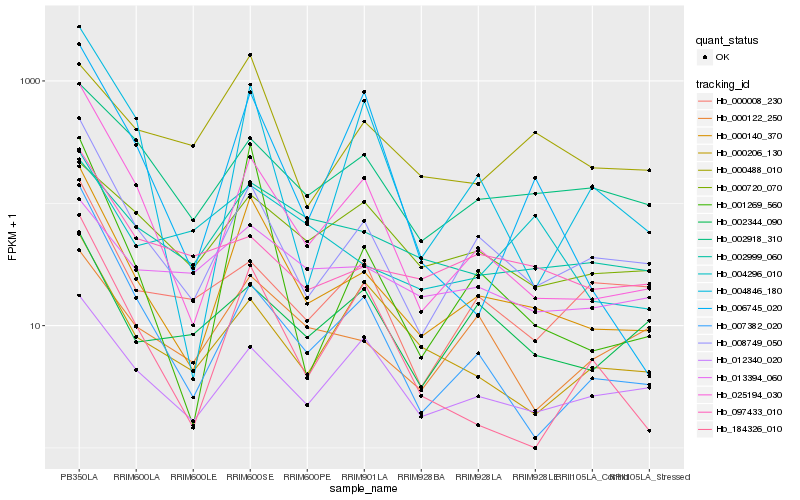
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_370 |
0.0 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6 [Jatropha curcas] |
| 2 |
Hb_008749_050 |
0.1475537814 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C4H3.01 [Jatropha curcas] |
| 3 |
Hb_025194_030 |
0.1749461588 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 4 |
Hb_001269_560 |
0.1783876281 |
- |
- |
heat-shock protein 70 [Hevea brasiliensis] |
| 5 |
Hb_004846_180 |
0.2087549301 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 6 |
Hb_012340_020 |
0.213786776 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 7 |
Hb_002918_310 |
0.2169198729 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 8 |
Hb_000122_250 |
0.2224597704 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 9 |
Hb_002999_060 |
0.2250124927 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 10 |
Hb_184326_010 |
0.2279008294 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 11 |
Hb_000488_010 |
0.2325308859 |
- |
- |
18.1 kDa class I heat shock protein [Jatropha curcas] |
| 12 |
Hb_000206_130 |
0.2364909434 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_002344_090 |
0.239939794 |
- |
- |
PREDICTED: uncharacterized protein LOC105637976 [Jatropha curcas] |
| 14 |
Hb_006745_020 |
0.2417717335 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 15 |
Hb_007382_020 |
0.2439867179 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 16 |
Hb_013394_060 |
0.2457360581 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 17 |
Hb_097433_010 |
0.2479336906 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TDX [Jatropha curcas] |
| 18 |
Hb_000720_070 |
0.2479806195 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 19 |
Hb_004296_010 |
0.2484824068 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 20 |
Hb_000008_230 |
0.2486820851 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |