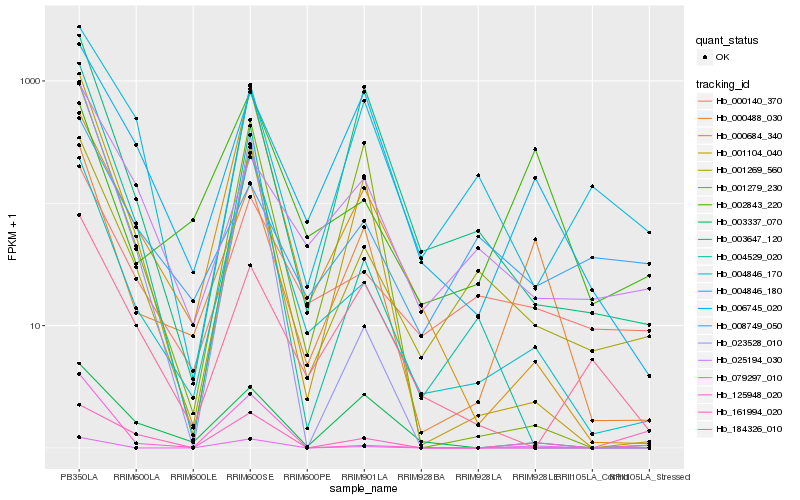
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_560 |
0.0 |
- |
- |
heat-shock protein 70 [Hevea brasiliensis] |
| 2 |
Hb_004846_170 |
0.1505463253 |
- |
- |
heat-shock protein 70 [Hevea brasiliensis] |
| 3 |
Hb_000140_370 |
0.1783876281 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6 [Jatropha curcas] |
| 4 |
Hb_000684_340 |
0.2061451728 |
- |
- |
PREDICTED: L-ascorbate peroxidase 2, cytosolic [Pyrus x bretschneideri] |
| 5 |
Hb_125948_020 |
0.2210862299 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 6 |
Hb_004846_180 |
0.2414208881 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 7 |
Hb_003647_120 |
0.2563607449 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 8 |
Hb_000488_030 |
0.2597723897 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 9 |
Hb_001279_230 |
0.2607561894 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 10 |
Hb_025194_030 |
0.2640132885 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 11 |
Hb_079297_010 |
0.2641477281 |
- |
- |
hypothetical protein VITISV_012409 [Vitis vinifera] |
| 12 |
Hb_184326_010 |
0.2649571353 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 13 |
Hb_008749_050 |
0.2671193314 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C4H3.01 [Jatropha curcas] |
| 14 |
Hb_003337_070 |
0.2707785032 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 15 |
Hb_004529_020 |
0.2709693507 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 16 |
Hb_002843_220 |
0.2856090906 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic [Jatropha curcas] |
| 17 |
Hb_006745_020 |
0.2900272511 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 18 |
Hb_023528_010 |
0.2904104237 |
- |
- |
hypothetical protein POPTR_0013s01580g [Populus trichocarpa] |
| 19 |
Hb_001104_040 |
0.2950593492 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 20 |
Hb_161994_020 |
0.2957052553 |
- |
- |
- |