| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003637_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_086484_010 |
0.0205484253 |
- |
- |
PREDICTED: uncharacterized protein LOC101259761 [Solanum lycopersicum] |
| 3 |
Hb_102355_010 |
0.0247261651 |
- |
- |
PREDICTED: uncharacterized protein LOC104094591, partial [Nicotiana tomentosiformis] |
| 4 |
Hb_002708_010 |
0.0456243603 |
- |
- |
- |
| 5 |
Hb_185830_100 |
0.0472942646 |
- |
- |
- |
| 6 |
Hb_001863_210 |
0.0488009509 |
- |
- |
- |
| 7 |
Hb_006387_010 |
0.0573239453 |
- |
- |
PREDICTED: uncharacterized protein LOC105649124 [Jatropha curcas] |
| 8 |
Hb_049827_010 |
0.0609245463 |
- |
- |
PREDICTED: uncharacterized protein LOC105762110 [Gossypium raimondii] |
| 9 |
Hb_003392_010 |
0.0639895493 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 10 |
Hb_012264_040 |
0.0867859012 |
- |
- |
- |
| 11 |
Hb_135941_010 |
0.0984989931 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 12 |
Hb_133862_010 |
0.1018480984 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 13 |
Hb_001066_020 |
0.1176183036 |
- |
- |
PREDICTED: uncharacterized protein LOC104908874, partial [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_005432_050 |
0.140109908 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |
| 15 |
Hb_007943_010 |
0.1486567119 |
- |
- |
- |
| 16 |
Hb_008424_010 |
0.1735647408 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_000016_010 |
0.1748427808 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 18 |
Hb_038190_040 |
0.2172483859 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 19 |
Hb_125948_020 |
0.2225166921 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 20 |
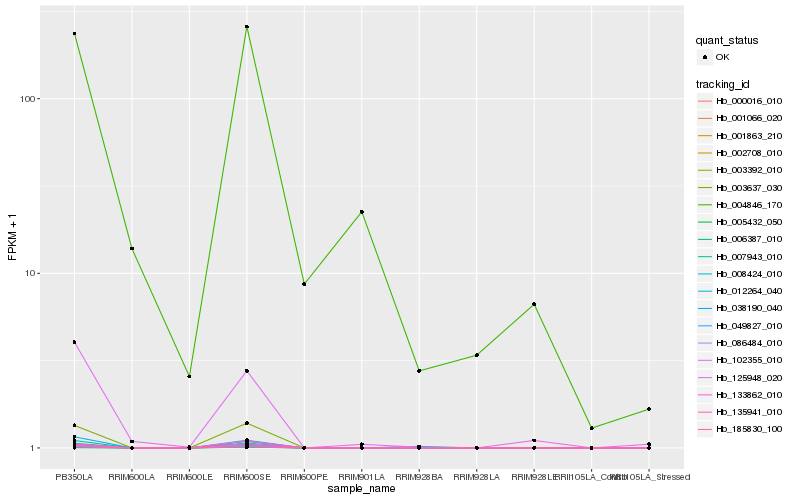
Hb_004846_170 |
0.2271238031 |
- |
- |
heat-shock protein 70 [Hevea brasiliensis] |