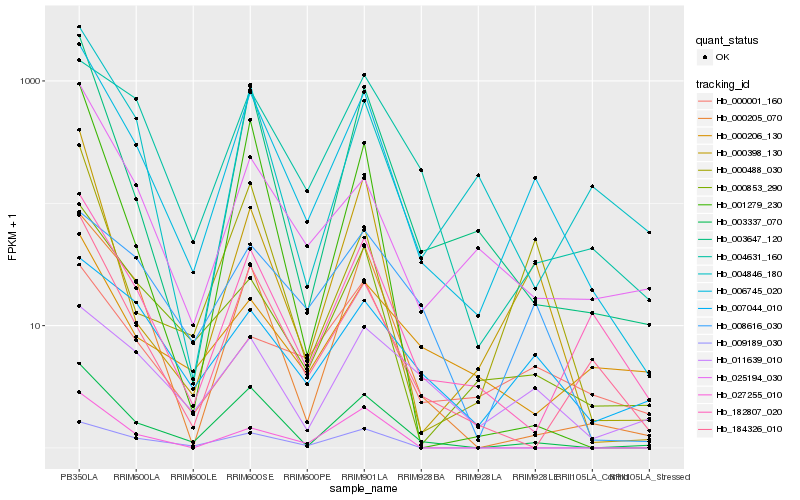
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006745_020 |
0.0 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 2 |
Hb_003337_070 |
0.1356440333 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 3 |
Hb_000853_290 |
0.1658412163 |
- |
- |
PREDICTED: uncharacterized protein LOC105642585 [Jatropha curcas] |
| 4 |
Hb_000205_070 |
0.1747708012 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_184326_010 |
0.1771311535 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 6 |
Hb_003647_120 |
0.1788059414 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 7 |
Hb_000398_130 |
0.178891125 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 8 |
Hb_182807_020 |
0.1845884172 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 9 |
Hb_000488_030 |
0.1880425424 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 10 |
Hb_007044_010 |
0.1950332684 |
- |
- |
- |
| 11 |
Hb_025194_030 |
0.1950873262 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 12 |
Hb_009189_030 |
0.2061381806 |
- |
- |
hypothetical protein PRUPE_ppa023969mg, partial [Prunus persica] |
| 13 |
Hb_008616_030 |
0.2093313031 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004846_180 |
0.2096942046 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 15 |
Hb_027255_010 |
0.2105533185 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004631_160 |
0.2108987943 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 17 |
Hb_001279_230 |
0.21436748 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 18 |
Hb_000001_160 |
0.2166449842 |
- |
- |
PREDICTED: probable glutamate carboxypeptidase 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_011639_010 |
0.2240594668 |
- |
- |
- |
| 20 |
Hb_000206_130 |
0.2354296425 |
- |
- |
unnamed protein product [Vitis vinifera] |