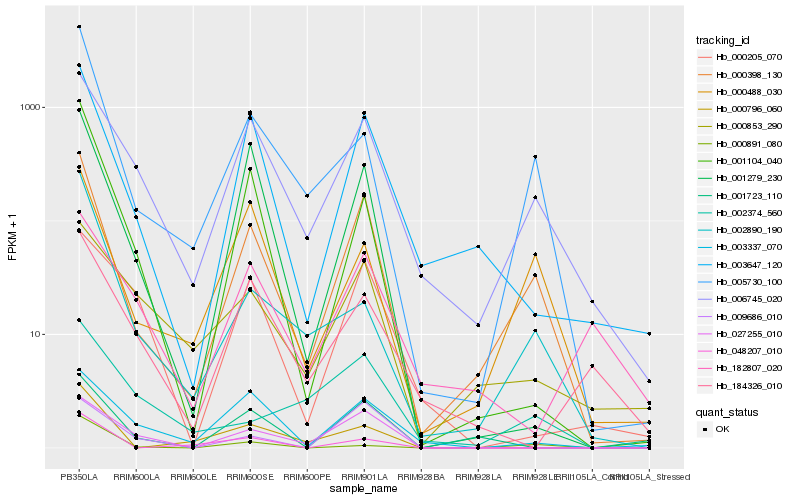
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000398_130 |
0.0 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 2 |
Hb_006745_020 |
0.178891125 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 3 |
Hb_003647_120 |
0.1792959684 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 4 |
Hb_000488_030 |
0.2048190539 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 5 |
Hb_005730_100 |
0.207503787 |
- |
- |
PREDICTED: 18.2 kDa class I heat shock protein-like [Gossypium raimondii] |
| 6 |
Hb_001279_230 |
0.2196966699 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 7 |
Hb_027255_010 |
0.2295536101 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001723_110 |
0.2331862881 |
- |
- |
PREDICTED: solute carrier family 25 member 44-like isoform X2 [Malus domestica] |
| 9 |
Hb_001104_040 |
0.2356596587 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 10 |
Hb_003337_070 |
0.2366791255 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 11 |
Hb_000853_290 |
0.239274822 |
- |
- |
PREDICTED: uncharacterized protein LOC105642585 [Jatropha curcas] |
| 12 |
Hb_000796_060 |
0.2513296577 |
- |
- |
Glycogenin-2 [Aegilops tauschii] |
| 13 |
Hb_009686_010 |
0.2549692946 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |
| 14 |
Hb_048207_010 |
0.2575728184 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 15 |
Hb_000205_070 |
0.2643114348 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_184326_010 |
0.2653692758 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 17 |
Hb_182807_020 |
0.2691223001 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 18 |
Hb_000891_080 |
0.2719305383 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Populus euphratica] |
| 19 |
Hb_002374_560 |
0.272493434 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH11-like [Populus euphratica] |
| 20 |
Hb_002890_190 |
0.2753684985 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |