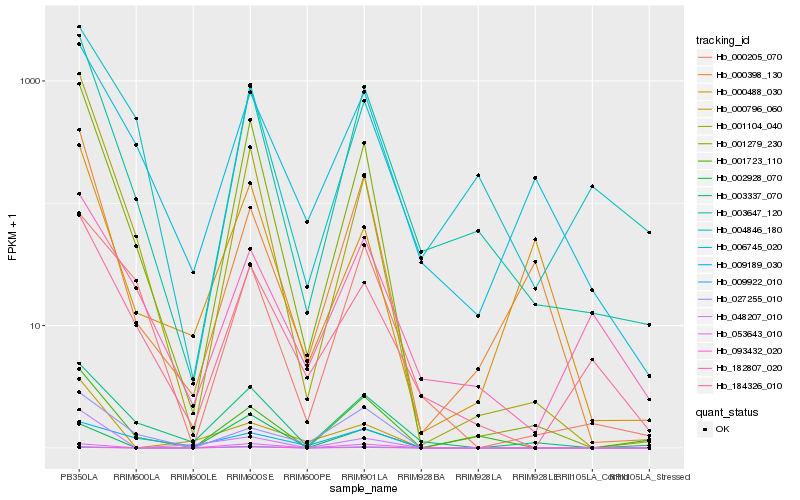
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001279_230 |
0.0 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 2 |
Hb_003647_120 |
0.1333330367 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 3 |
Hb_001104_040 |
0.1527172361 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 4 |
Hb_003337_070 |
0.1794476994 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 5 |
Hb_184326_010 |
0.1957389624 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 6 |
Hb_001723_110 |
0.2079579551 |
- |
- |
PREDICTED: solute carrier family 25 member 44-like isoform X2 [Malus domestica] |
| 7 |
Hb_006745_020 |
0.21436748 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 8 |
Hb_053643_010 |
0.2169806202 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 9 |
Hb_000205_070 |
0.2189508531 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000398_130 |
0.2196966699 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 11 |
Hb_027255_010 |
0.2258751272 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_048207_010 |
0.2290695037 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 13 |
Hb_000488_030 |
0.2353188412 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 14 |
Hb_000796_060 |
0.235398727 |
- |
- |
Glycogenin-2 [Aegilops tauschii] |
| 15 |
Hb_009922_010 |
0.237596331 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 16 |
Hb_002928_070 |
0.2418705659 |
- |
- |
- |
| 17 |
Hb_182807_020 |
0.2469474935 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 18 |
Hb_009189_030 |
0.2478064631 |
- |
- |
hypothetical protein PRUPE_ppa023969mg, partial [Prunus persica] |
| 19 |
Hb_004846_180 |
0.2497446601 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 20 |
Hb_093432_020 |
0.2573748951 |
- |
- |
PREDICTED: QWRF motif-containing protein 3 [Jatropha curcas] |