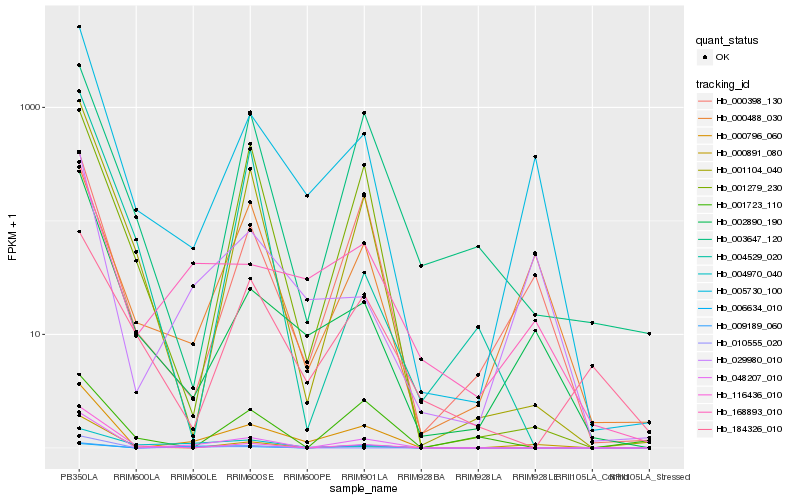
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048207_010 |
0.0 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 2 |
Hb_000796_060 |
0.1255162918 |
- |
- |
Glycogenin-2 [Aegilops tauschii] |
| 3 |
Hb_001104_040 |
0.1960428142 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 4 |
Hb_001279_230 |
0.2290695037 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 5 |
Hb_006634_010 |
0.2313552052 |
- |
- |
- |
| 6 |
Hb_009189_060 |
0.2323705475 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005730_100 |
0.2446346136 |
- |
- |
PREDICTED: 18.2 kDa class I heat shock protein-like [Gossypium raimondii] |
| 8 |
Hb_003647_120 |
0.2480882387 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 9 |
Hb_000398_130 |
0.2575728184 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 10 |
Hb_168893_010 |
0.2634883474 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 11 |
Hb_116436_010 |
0.2646972185 |
- |
- |
PREDICTED: uncharacterized protein LOC105766967 [Gossypium raimondii] |
| 12 |
Hb_010555_020 |
0.2696189122 |
- |
- |
hypothetical protein JCGZ_06771 [Jatropha curcas] |
| 13 |
Hb_002890_190 |
0.2815686191 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 14 |
Hb_000891_080 |
0.2822897658 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Populus euphratica] |
| 15 |
Hb_004529_020 |
0.2840517302 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 16 |
Hb_000488_030 |
0.2862048149 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 17 |
Hb_029980_010 |
0.2888870896 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 18 |
Hb_184326_010 |
0.2910623743 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 19 |
Hb_004970_040 |
0.2955535174 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 20 |
Hb_001723_110 |
0.2967192715 |
- |
- |
PREDICTED: solute carrier family 25 member 44-like isoform X2 [Malus domestica] |