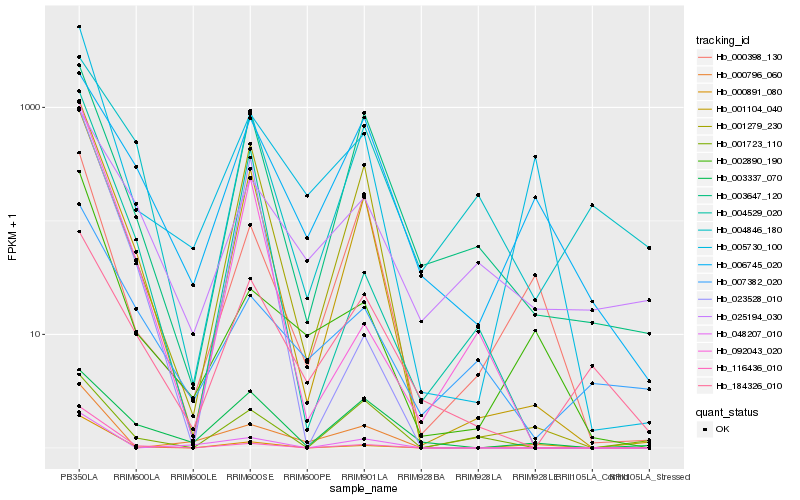
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001104_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 2 |
Hb_001279_230 |
0.1527172361 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 3 |
Hb_004529_020 |
0.1677736695 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 4 |
Hb_116436_010 |
0.1831210613 |
- |
- |
PREDICTED: uncharacterized protein LOC105766967 [Gossypium raimondii] |
| 5 |
Hb_003647_120 |
0.1850915494 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 6 |
Hb_092043_020 |
0.195888542 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 7 |
Hb_048207_010 |
0.1960428142 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 8 |
Hb_005730_100 |
0.2022237485 |
- |
- |
PREDICTED: 18.2 kDa class I heat shock protein-like [Gossypium raimondii] |
| 9 |
Hb_023528_010 |
0.2083580206 |
- |
- |
hypothetical protein POPTR_0013s01580g [Populus trichocarpa] |
| 10 |
Hb_184326_010 |
0.2114357062 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 11 |
Hb_000891_080 |
0.212326051 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Populus euphratica] |
| 12 |
Hb_002890_190 |
0.2212262132 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 13 |
Hb_000796_060 |
0.2212422731 |
- |
- |
Glycogenin-2 [Aegilops tauschii] |
| 14 |
Hb_007382_020 |
0.2272582082 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 15 |
Hb_025194_030 |
0.2344799304 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 16 |
Hb_000398_130 |
0.2356596587 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 17 |
Hb_004846_180 |
0.2401289013 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 18 |
Hb_001723_110 |
0.2474655387 |
- |
- |
PREDICTED: solute carrier family 25 member 44-like isoform X2 [Malus domestica] |
| 19 |
Hb_003337_070 |
0.2480206427 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 20 |
Hb_006745_020 |
0.2516354053 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |