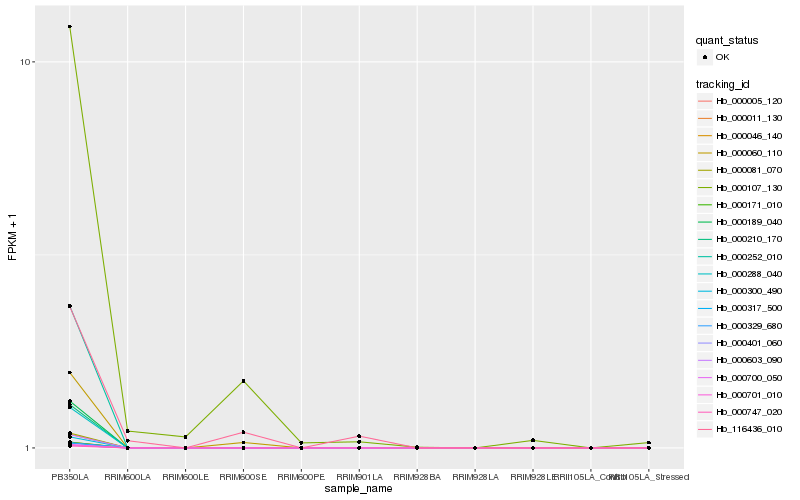
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_130 |
0.0 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 2 |
Hb_000060_110 |
0.1191764043 |
- |
- |
PREDICTED: uncharacterized protein LOC105649714 [Jatropha curcas] |
| 3 |
Hb_116436_010 |
0.1771425283 |
- |
- |
PREDICTED: uncharacterized protein LOC105766967 [Gossypium raimondii] |
| 4 |
Hb_000005_120 |
0.1854838127 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 5 |
Hb_000011_130 |
0.1854838127 |
- |
- |
lipase [Ricinus communis] |
| 6 |
Hb_000046_140 |
0.1854838127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000081_070 |
0.1854838127 |
- |
- |
- |
| 8 |
Hb_000171_010 |
0.1854838127 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 9 |
Hb_000189_040 |
0.1854838127 |
- |
- |
hypothetical protein [Trichoderma aureoviride] |
| 10 |
Hb_000210_170 |
0.1854838127 |
- |
- |
- |
| 11 |
Hb_000252_010 |
0.1854838127 |
- |
- |
- |
| 12 |
Hb_000288_040 |
0.1854838127 |
- |
- |
- |
| 13 |
Hb_000300_490 |
0.1854838127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000317_500 |
0.1854838127 |
- |
- |
- |
| 15 |
Hb_000329_680 |
0.1854838127 |
- |
- |
hypothetical protein JCGZ_14616 [Jatropha curcas] |
| 16 |
Hb_000401_060 |
0.1854838127 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 17 |
Hb_000603_090 |
0.1854838127 |
- |
- |
- |
| 18 |
Hb_000700_050 |
0.1854838127 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like isoform X5 [Jatropha curcas] |
| 19 |
Hb_000701_010 |
0.1854838127 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 20 |
Hb_000747_020 |
0.1854838127 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |