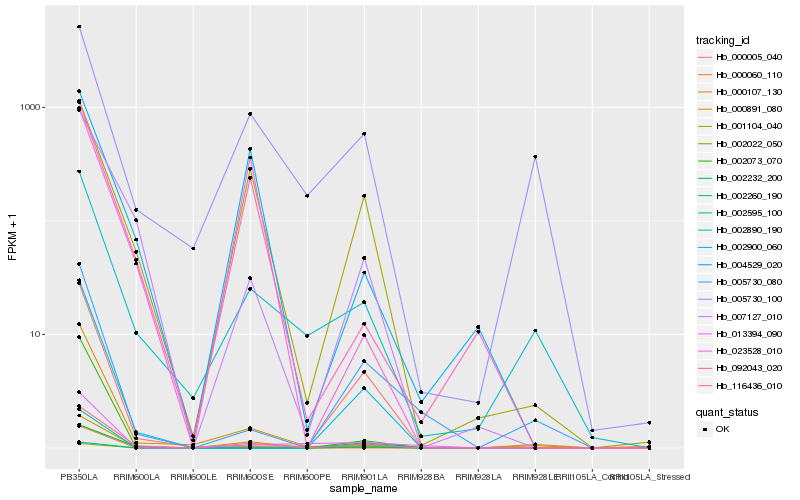
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116436_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105766967 [Gossypium raimondii] |
| 2 |
Hb_007127_010 |
0.1342273244 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 3 |
Hb_000107_130 |
0.1771425283 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 4 |
Hb_002890_190 |
0.1795203191 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 5 |
Hb_001104_040 |
0.1831210613 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 6 |
Hb_092043_020 |
0.1845740168 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 7 |
Hb_002900_060 |
0.193233944 |
- |
- |
CI small heat shock protein 2 [Prunus salicina] |
| 8 |
Hb_000891_080 |
0.1954151149 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Populus euphratica] |
| 9 |
Hb_000060_110 |
0.1992854637 |
- |
- |
PREDICTED: uncharacterized protein LOC105649714 [Jatropha curcas] |
| 10 |
Hb_000005_040 |
0.2133242836 |
- |
- |
PREDICTED: histone H2B.2 [Jatropha curcas] |
| 11 |
Hb_004529_020 |
0.2162375579 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 12 |
Hb_005730_080 |
0.2177380596 |
- |
- |
PREDICTED: 17.6 kDa class I heat shock protein-like [Solanum lycopersicum] |
| 13 |
Hb_002595_100 |
0.2199945948 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 14 |
Hb_013394_090 |
0.2213818752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002073_070 |
0.2322771985 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 16 |
Hb_005730_100 |
0.2378669514 |
- |
- |
PREDICTED: 18.2 kDa class I heat shock protein-like [Gossypium raimondii] |
| 17 |
Hb_002232_200 |
0.2429190725 |
- |
- |
PREDICTED: sugar carrier protein C-like [Jatropha curcas] |
| 18 |
Hb_002260_190 |
0.2451522587 |
- |
- |
- |
| 19 |
Hb_002022_050 |
0.2486020038 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like isoform X1 [Populus euphratica] |
| 20 |
Hb_023528_010 |
0.2524933092 |
- |
- |
hypothetical protein POPTR_0013s01580g [Populus trichocarpa] |