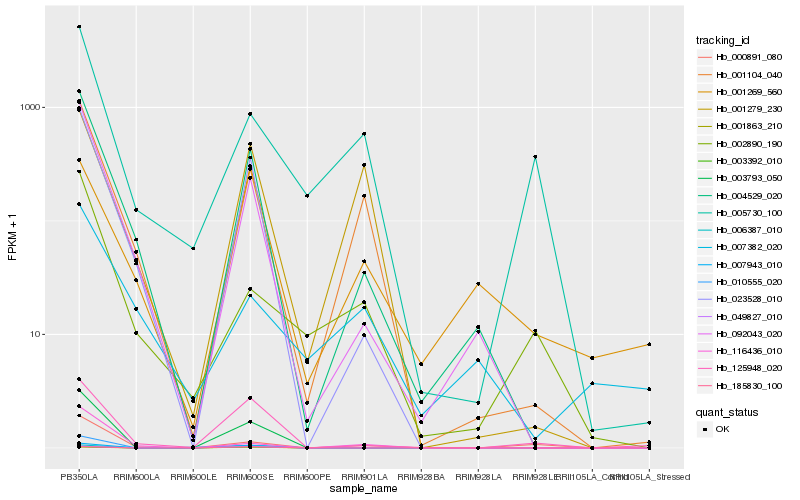
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004529_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 2 |
Hb_092043_020 |
0.0721354982 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 3 |
Hb_023528_010 |
0.0806040021 |
- |
- |
hypothetical protein POPTR_0013s01580g [Populus trichocarpa] |
| 4 |
Hb_001104_040 |
0.1677736695 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 5 |
Hb_003793_050 |
0.1739968415 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 6 |
Hb_125948_020 |
0.189327118 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 7 |
Hb_007943_010 |
0.205032482 |
- |
- |
- |
| 8 |
Hb_116436_010 |
0.2162375579 |
- |
- |
PREDICTED: uncharacterized protein LOC105766967 [Gossypium raimondii] |
| 9 |
Hb_000891_080 |
0.2344281506 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Populus euphratica] |
| 10 |
Hb_010555_020 |
0.2571238652 |
- |
- |
hypothetical protein JCGZ_06771 [Jatropha curcas] |
| 11 |
Hb_003392_010 |
0.2585959502 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 12 |
Hb_002890_190 |
0.2604024288 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 13 |
Hb_049827_010 |
0.260804186 |
- |
- |
PREDICTED: uncharacterized protein LOC105762110 [Gossypium raimondii] |
| 14 |
Hb_001279_230 |
0.2632772274 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 15 |
Hb_006387_010 |
0.2634160646 |
- |
- |
PREDICTED: uncharacterized protein LOC105649124 [Jatropha curcas] |
| 16 |
Hb_007382_020 |
0.2650266187 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 17 |
Hb_005730_100 |
0.2657508736 |
- |
- |
PREDICTED: 18.2 kDa class I heat shock protein-like [Gossypium raimondii] |
| 18 |
Hb_001863_210 |
0.2696715925 |
- |
- |
- |
| 19 |
Hb_185830_100 |
0.2707876608 |
- |
- |
- |
| 20 |
Hb_001269_560 |
0.2709693507 |
- |
- |
heat-shock protein 70 [Hevea brasiliensis] |