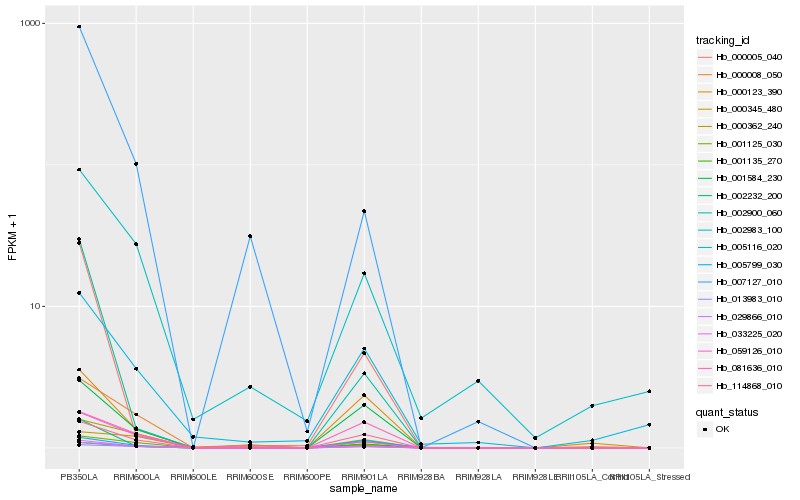
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033225_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_059126_010 |
0.0759775182 |
- |
- |
- |
| 3 |
Hb_029866_010 |
0.0984245315 |
- |
- |
- |
| 4 |
Hb_001125_030 |
0.1192085116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_013983_010 |
0.1533308212 |
- |
- |
hypothetical protein PRUPE_ppa016763mg, partial [Prunus persica] |
| 6 |
Hb_001584_230 |
0.1772343728 |
- |
- |
invertase/pectin methylesterase inhibitor family protein [Populus trichocarpa] |
| 7 |
Hb_002983_100 |
0.1788086854 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF084 [Jatropha curcas] |
| 8 |
Hb_002232_200 |
0.1859895974 |
- |
- |
PREDICTED: sugar carrier protein C-like [Jatropha curcas] |
| 9 |
Hb_007127_010 |
0.2021944148 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 10 |
Hb_005116_020 |
0.2026841212 |
- |
- |
PREDICTED: DNA repair helicase UVH6 isoform X2 [Sesamum indicum] |
| 11 |
Hb_114868_010 |
0.2081759982 |
- |
- |
- |
| 12 |
Hb_081636_010 |
0.2116809448 |
- |
- |
- |
| 13 |
Hb_000123_390 |
0.2195053359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000008_050 |
0.2222328767 |
- |
- |
Phenylalanine ammonia-lyase, putative [Ricinus communis] |
| 15 |
Hb_000345_480 |
0.227587986 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_005799_030 |
0.2315820882 |
- |
- |
PREDICTED: uncharacterized protein LOC103926623 [Pyrus x bretschneideri] |
| 17 |
Hb_001135_270 |
0.237662494 |
- |
- |
- |
| 18 |
Hb_002900_060 |
0.2533031132 |
- |
- |
CI small heat shock protein 2 [Prunus salicina] |
| 19 |
Hb_000005_040 |
0.2544953808 |
- |
- |
PREDICTED: histone H2B.2 [Jatropha curcas] |
| 20 |
Hb_000362_240 |
0.2557947365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |