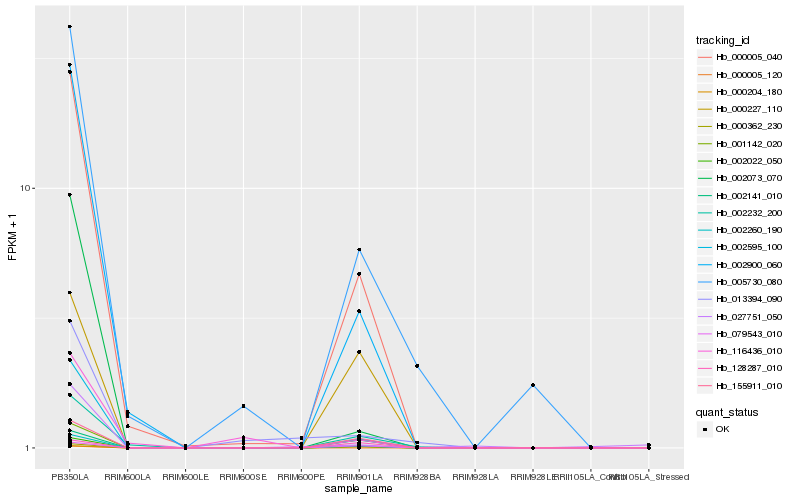
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002260_190 |
0.0 |
- |
- |
- |
| 2 |
Hb_002022_050 |
0.0078589379 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_000005_040 |
0.0713616588 |
- |
- |
PREDICTED: histone H2B.2 [Jatropha curcas] |
| 4 |
Hb_002595_100 |
0.0914148366 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 5 |
Hb_002900_060 |
0.1064820104 |
- |
- |
CI small heat shock protein 2 [Prunus salicina] |
| 6 |
Hb_001142_020 |
0.1189289825 |
- |
- |
- |
| 7 |
Hb_155911_010 |
0.1247457189 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 8 |
Hb_000362_230 |
0.1367750741 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_079543_010 |
0.1517866892 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 10 |
Hb_002232_200 |
0.1636570717 |
- |
- |
PREDICTED: sugar carrier protein C-like [Jatropha curcas] |
| 11 |
Hb_005730_080 |
0.1680614406 |
- |
- |
PREDICTED: 17.6 kDa class I heat shock protein-like [Solanum lycopersicum] |
| 12 |
Hb_002141_010 |
0.1798136518 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_002073_070 |
0.1882350879 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 14 |
Hb_000227_110 |
0.1913077328 |
- |
- |
hypothetical protein CICLE_v10010500mg, partial [Citrus clementina] |
| 15 |
Hb_000204_180 |
0.218468377 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 16 |
Hb_013394_090 |
0.2383521361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_116436_010 |
0.2451522587 |
- |
- |
PREDICTED: uncharacterized protein LOC105766967 [Gossypium raimondii] |
| 18 |
Hb_027751_050 |
0.252051502 |
- |
- |
hypothetical protein JCGZ_05887 [Jatropha curcas] |
| 19 |
Hb_128287_010 |
0.2586170025 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 20 |
Hb_000005_120 |
0.2587477189 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |