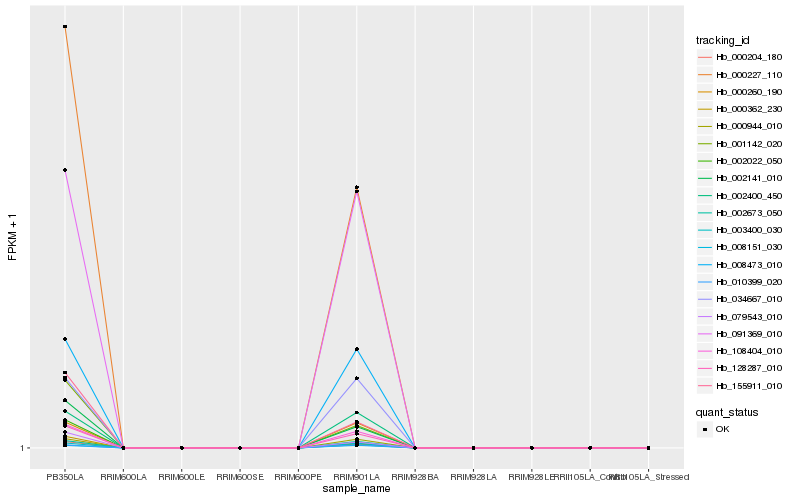
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_110 |
0.0 |
- |
- |
hypothetical protein CICLE_v10010500mg, partial [Citrus clementina] |
| 2 |
Hb_002141_010 |
0.0117365119 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000204_180 |
0.0278192267 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 4 |
Hb_079543_010 |
0.0402545039 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 5 |
Hb_000362_230 |
0.0554655466 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_155911_010 |
0.0676198208 |
- |
- |
hypothetical protein JCGZ_26951 [Jatropha curcas] |
| 7 |
Hb_128287_010 |
0.0691382349 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 8 |
Hb_001142_020 |
0.0734852337 |
- |
- |
- |
| 9 |
Hb_108404_010 |
0.087854516 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 10 |
Hb_002673_050 |
0.1361045566 |
- |
- |
- |
| 11 |
Hb_091369_010 |
0.1381212931 |
- |
- |
- |
| 12 |
Hb_008473_010 |
0.139097605 |
- |
- |
PREDICTED: laccase-14-like [Jatropha curcas] |
| 13 |
Hb_000944_010 |
0.1496957161 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 14 |
Hb_003400_030 |
0.1497932019 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 15 |
Hb_002400_450 |
0.1544807667 |
- |
- |
PREDICTED: protein MEMO1 isoform X2 [Sesamum indicum] |
| 16 |
Hb_000260_190 |
0.1569018799 |
- |
- |
- |
| 17 |
Hb_034667_010 |
0.1610238195 |
- |
- |
- |
| 18 |
Hb_008151_030 |
0.1674634049 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_010399_020 |
0.1713278893 |
- |
- |
PREDICTED: uncharacterized protein LOC104094989 [Nicotiana tomentosiformis] |
| 20 |
Hb_002022_050 |
0.1836702746 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like isoform X1 [Populus euphratica] |