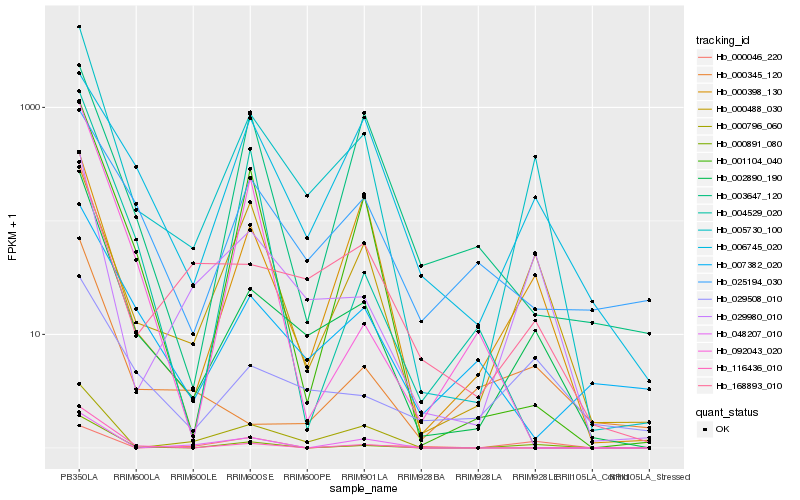
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005730_100 |
0.0 |
- |
- |
PREDICTED: 18.2 kDa class I heat shock protein-like [Gossypium raimondii] |
| 2 |
Hb_002890_190 |
0.1114687766 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 3 |
Hb_000891_080 |
0.1447605033 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Populus euphratica] |
| 4 |
Hb_029980_010 |
0.1579480864 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 5 |
Hb_000488_030 |
0.1948911195 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 6 |
Hb_001104_040 |
0.2022237485 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 7 |
Hb_000398_130 |
0.207503787 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 8 |
Hb_168893_010 |
0.2157502941 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |
| 9 |
Hb_000796_060 |
0.2201165794 |
- |
- |
Glycogenin-2 [Aegilops tauschii] |
| 10 |
Hb_029508_010 |
0.2348876006 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_116436_010 |
0.2378669514 |
- |
- |
PREDICTED: uncharacterized protein LOC105766967 [Gossypium raimondii] |
| 12 |
Hb_006745_020 |
0.2435620277 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 13 |
Hb_025194_030 |
0.2444159528 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 14 |
Hb_048207_010 |
0.2446346136 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 15 |
Hb_007382_020 |
0.2477066668 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 16 |
Hb_004529_020 |
0.2657508736 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 17 |
Hb_092043_020 |
0.2681140137 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 18 |
Hb_000046_220 |
0.2715974802 |
- |
- |
- |
| 19 |
Hb_003647_120 |
0.2750392293 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 20 |
Hb_000345_120 |
0.2750586739 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 37a [Jatropha curcas] |