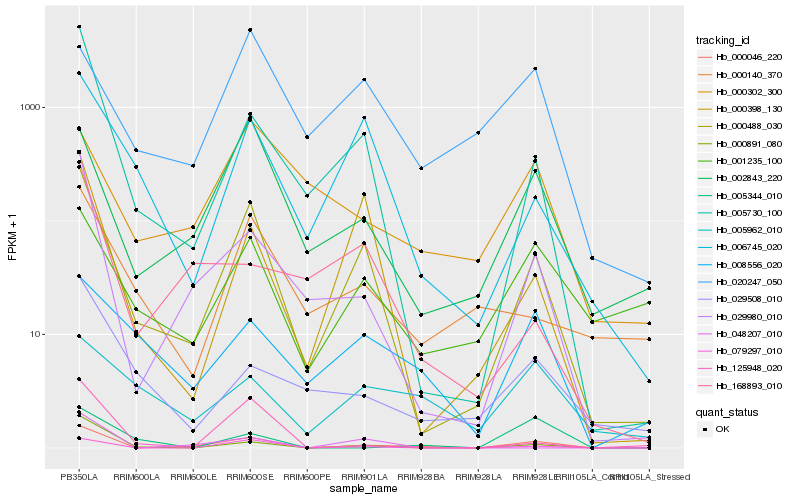
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_220 |
0.0 |
- |
- |
- |
| 2 |
Hb_000488_030 |
0.2047980043 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 3 |
Hb_029980_010 |
0.2196715066 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 4 |
Hb_079297_010 |
0.2305911831 |
- |
- |
hypothetical protein VITISV_012409 [Vitis vinifera] |
| 5 |
Hb_005730_100 |
0.2715974802 |
- |
- |
PREDICTED: 18.2 kDa class I heat shock protein-like [Gossypium raimondii] |
| 6 |
Hb_000891_080 |
0.2772257417 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Populus euphratica] |
| 7 |
Hb_002843_220 |
0.2774866044 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic [Jatropha curcas] |
| 8 |
Hb_125948_020 |
0.3009588825 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 9 |
Hb_001235_100 |
0.3144396137 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 10 |
Hb_008556_020 |
0.3172126846 |
- |
- |
- |
| 11 |
Hb_048207_010 |
0.3227124486 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 12 |
Hb_000398_130 |
0.326854797 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein-like [Jatropha curcas] |
| 13 |
Hb_029508_010 |
0.3312431614 |
transcription factor |
TF Family: HSF |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_006745_020 |
0.3327149592 |
- |
- |
PREDICTED: 22.0 kDa class IV heat shock protein [Jatropha curcas] |
| 15 |
Hb_000302_300 |
0.3371803788 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 16 |
Hb_005344_010 |
0.340707534 |
- |
- |
PREDICTED: uncharacterized protein LOC102597679 [Solanum tuberosum] |
| 17 |
Hb_000140_370 |
0.3410321608 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6 [Jatropha curcas] |
| 18 |
Hb_005962_010 |
0.3428220261 |
- |
- |
- |
| 19 |
Hb_020247_050 |
0.3437799496 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 20 |
Hb_168893_010 |
0.3446662135 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Nelumbo nucifera] |