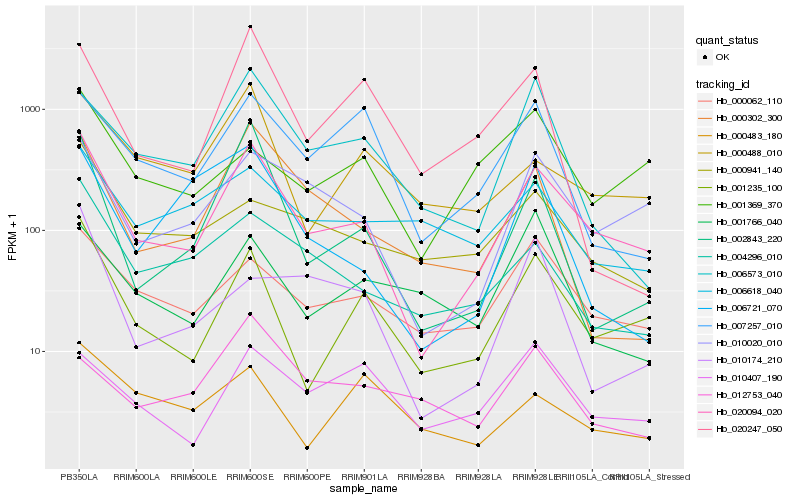
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020094_020 |
0.0 |
- |
- |
PREDICTED: 17.1 kDa class II heat shock protein-like [Pyrus x bretschneideri] |
| 2 |
Hb_001235_100 |
0.1428813767 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 3 |
Hb_010020_010 |
0.1579277823 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 4 |
Hb_002843_220 |
0.1844067857 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000062_110 |
0.1858315379 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 6 |
Hb_004296_010 |
0.1924189338 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 7 |
Hb_000302_300 |
0.1977611606 |
- |
- |
PREDICTED: heat shock protein 83 [Populus euphratica] |
| 8 |
Hb_000488_010 |
0.1984405435 |
- |
- |
18.1 kDa class I heat shock protein [Jatropha curcas] |
| 9 |
Hb_006573_010 |
0.2054627191 |
- |
- |
heat shock protein [Hevea brasiliensis] |
| 10 |
Hb_000941_140 |
0.2124774815 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 11 |
Hb_007257_010 |
0.2139406263 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 12 |
Hb_006618_040 |
0.2174852749 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 13 |
Hb_010407_190 |
0.2194050632 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 14 |
Hb_006721_070 |
0.225006732 |
- |
- |
PREDICTED: 70 kDa peptidyl-prolyl isomerase-like [Jatropha curcas] |
| 15 |
Hb_001369_370 |
0.2258498433 |
- |
- |
PREDICTED: 23.6 kDa heat shock protein, mitochondrial [Jatropha curcas] |
| 16 |
Hb_020247_050 |
0.2290895175 |
- |
- |
PREDICTED: 18.1 kDa class I heat shock protein-like [Populus euphratica] |
| 17 |
Hb_001766_040 |
0.2310448737 |
- |
- |
chaperone clpb, putative [Ricinus communis] |
| 18 |
Hb_000483_180 |
0.2381062618 |
- |
- |
- |
| 19 |
Hb_010174_210 |
0.2389434529 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_012753_040 |
0.2434170195 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |