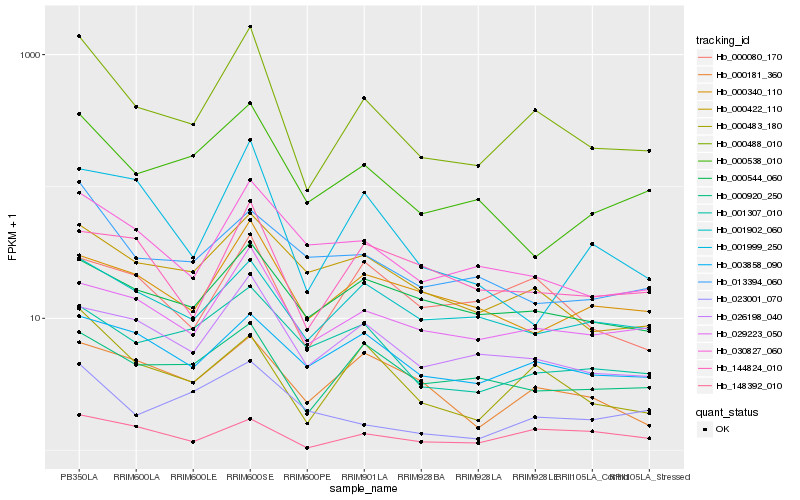
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000488_010 |
0.0 |
- |
- |
18.1 kDa class I heat shock protein [Jatropha curcas] |
| 2 |
Hb_000483_180 |
0.15112908 |
- |
- |
- |
| 3 |
Hb_026198_040 |
0.1609758257 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 4 |
Hb_029223_050 |
0.1675947007 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 5 |
Hb_030827_060 |
0.1692989789 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2b [Jatropha curcas] |
| 6 |
Hb_000538_010 |
0.1731067513 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000422_110 |
0.177171171 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 8 |
Hb_000920_250 |
0.1794292676 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein RCOM_0655500 [Ricinus communis] |
| 9 |
Hb_001307_010 |
0.1861412764 |
- |
- |
PREDICTED: uncharacterized protein LOC103939917 [Pyrus x bretschneideri] |
| 10 |
Hb_000544_060 |
0.1888599084 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 11 |
Hb_148392_010 |
0.1889322377 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_000080_170 |
0.1894453111 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_001902_060 |
0.1898139153 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |
| 14 |
Hb_144824_010 |
0.1917462481 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 15 |
Hb_000340_110 |
0.1918779163 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 16 |
Hb_001999_250 |
0.1925342041 |
- |
- |
PREDICTED: uncharacterized protein LOC105133073 isoform X1 [Populus euphratica] |
| 17 |
Hb_000181_360 |
0.1934162702 |
- |
- |
PREDICTED: uncharacterized protein LOC105644273 [Jatropha curcas] |
| 18 |
Hb_023001_070 |
0.1950836694 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A-like [Eucalyptus grandis] |
| 19 |
Hb_003858_090 |
0.1950873991 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 20 |
Hb_013394_060 |
0.195981635 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |