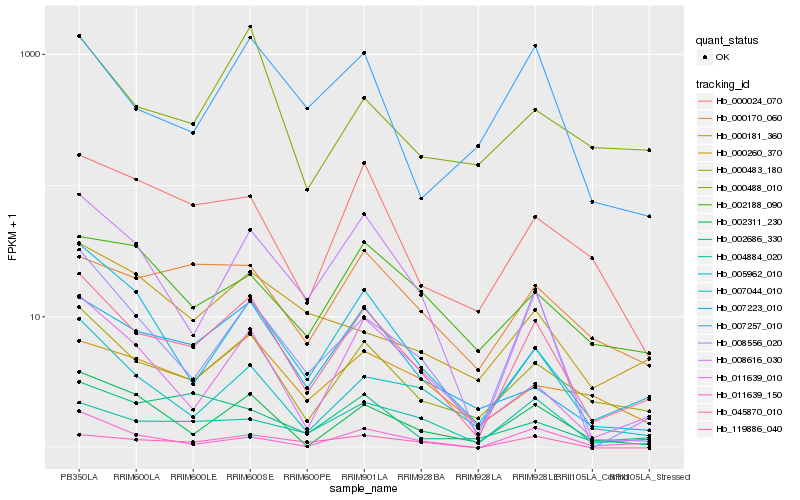
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_119886_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000483_180 |
0.1146359734 |
- |
- |
- |
| 3 |
Hb_008556_020 |
0.1618675572 |
- |
- |
- |
| 4 |
Hb_002311_230 |
0.1659074906 |
- |
- |
- |
| 5 |
Hb_007223_010 |
0.1805106125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011639_010 |
0.1808911296 |
- |
- |
- |
| 7 |
Hb_005962_010 |
0.1814059527 |
- |
- |
- |
| 8 |
Hb_007044_010 |
0.1833684465 |
- |
- |
- |
| 9 |
Hb_011639_150 |
0.1858784879 |
- |
- |
- |
| 10 |
Hb_000024_070 |
0.1943241708 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_007257_010 |
0.1951286942 |
- |
- |
cytosolic class II low molecular weight heat shock protein [Prunus dulcis] |
| 12 |
Hb_000181_360 |
0.1957880737 |
- |
- |
PREDICTED: uncharacterized protein LOC105644273 [Jatropha curcas] |
| 13 |
Hb_000170_060 |
0.1993828579 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 14 |
Hb_045870_010 |
0.1994972493 |
- |
- |
- |
| 15 |
Hb_000488_010 |
0.1995102941 |
- |
- |
18.1 kDa class I heat shock protein [Jatropha curcas] |
| 16 |
Hb_008616_030 |
0.2007029234 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002188_090 |
0.2108552684 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X1 [Jatropha curcas] |
| 18 |
Hb_004884_020 |
0.2116578626 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 4 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000260_370 |
0.2117197734 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 20 |
Hb_002686_330 |
0.2126226085 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |