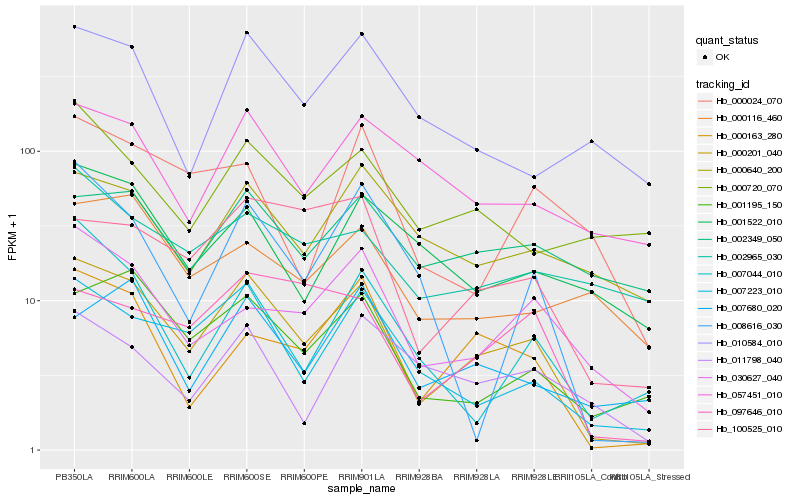
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000201_040 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 2 |
Hb_007223_010 |
0.162808102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001195_150 |
0.172155335 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 4 |
Hb_000163_280 |
0.1803205848 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 5 |
Hb_030627_040 |
0.1900809832 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 6 |
Hb_100525_010 |
0.1936448508 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 7 |
Hb_057451_010 |
0.2029553692 |
- |
- |
PREDICTED: uncharacterized protein LOC105635588 [Jatropha curcas] |
| 8 |
Hb_008616_030 |
0.2031383417 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 9 |
Hb_097646_010 |
0.2042477992 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 10 |
Hb_010584_010 |
0.2047577164 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 11 |
Hb_001522_010 |
0.2049069539 |
- |
- |
PREDICTED: nucleolar protein 14 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007680_020 |
0.2077133535 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000640_200 |
0.2081859461 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 14 |
Hb_007044_010 |
0.2088936078 |
- |
- |
- |
| 15 |
Hb_000720_070 |
0.212256597 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 16 |
Hb_000116_460 |
0.2133514836 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 17 |
Hb_002349_050 |
0.2142955893 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 18 |
Hb_000024_070 |
0.2144327844 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_011798_040 |
0.2149054953 |
- |
- |
hypothetical protein ZEAMMB73_778895 [Zea mays] |
| 20 |
Hb_002965_030 |
0.2156369636 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |