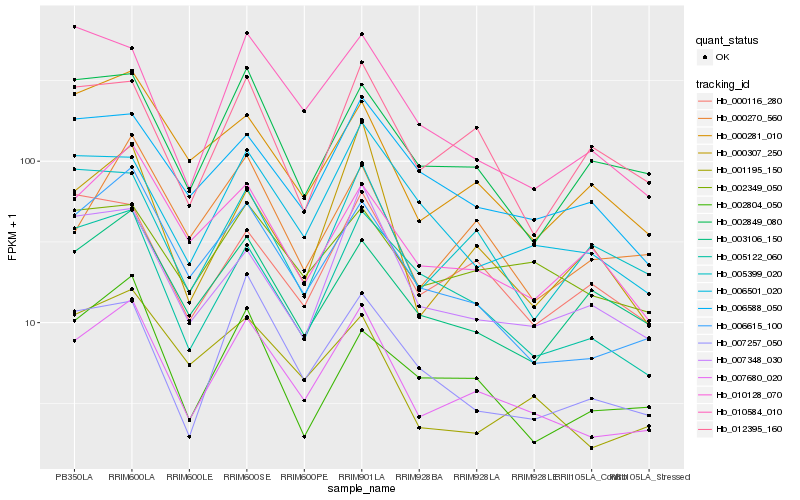
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007680_020 |
0.0 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006615_100 |
0.1103244313 |
- |
- |
PREDICTED: uncharacterized protein LOC105640683 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005122_060 |
0.1363983386 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_001195_150 |
0.1479218845 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 5 |
Hb_000307_250 |
0.1483823723 |
- |
- |
PREDICTED: probable nucleoredoxin 2 [Jatropha curcas] |
| 6 |
Hb_007257_050 |
0.1485815585 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_000281_010 |
0.1493546876 |
- |
- |
hypothetical protein JCGZ_16194 [Jatropha curcas] |
| 8 |
Hb_010128_070 |
0.1504523491 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_007348_030 |
0.1506016521 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X2 [Jatropha curcas] |
| 10 |
Hb_006501_020 |
0.1515693739 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 11 |
Hb_002849_080 |
0.1538380569 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog MMK2 [Jatropha curcas] |
| 12 |
Hb_000270_560 |
0.1554152743 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 13 |
Hb_002349_050 |
0.1565800641 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 14 |
Hb_006588_050 |
0.1575848254 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 15 |
Hb_002804_050 |
0.1586717551 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 16 |
Hb_012395_160 |
0.1594454687 |
- |
- |
PREDICTED: pre-mRNA-processing factor 19 homolog 1 [Jatropha curcas] |
| 17 |
Hb_010584_010 |
0.1595725298 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 18 |
Hb_005399_020 |
0.1600360615 |
- |
- |
hypothetical protein CISIN_1g022820mg [Citrus sinensis] |
| 19 |
Hb_000116_280 |
0.1603261751 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 20 |
Hb_003106_150 |
0.1615275117 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |