| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
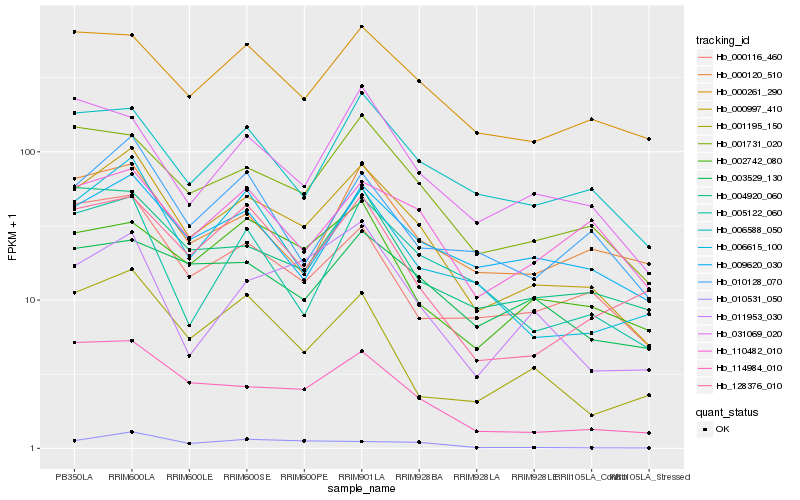
| 1 |
Hb_000997_410 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 2 |
Hb_001731_020 |
0.1183317321 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 3 |
Hb_114984_010 |
0.1267415495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_006615_100 |
0.1284999445 |
- |
- |
PREDICTED: uncharacterized protein LOC105640683 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000261_290 |
0.1373550606 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 6 |
Hb_006588_050 |
0.1373764304 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 7 |
Hb_009620_030 |
0.139582766 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |
| 8 |
Hb_004920_060 |
0.1450513549 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 9 |
Hb_010128_070 |
0.1467961126 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_000116_460 |
0.1477395338 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 11 |
Hb_128376_010 |
0.148177395 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_002742_080 |
0.1504084356 |
- |
- |
unknown [Populus trichocarpa] |
| 13 |
Hb_005122_060 |
0.1511630381 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_003529_130 |
0.1520621088 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_001195_150 |
0.1527616177 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 16 |
Hb_000120_510 |
0.1527991841 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_011953_030 |
0.1530638742 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0003s14450g [Populus trichocarpa] |
| 18 |
Hb_010531_050 |
0.1532164653 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 19 |
Hb_110482_010 |
0.1548539932 |
- |
- |
hypothetical protein JCGZ_16907 [Jatropha curcas] |
| 20 |
Hb_031069_020 |
0.1563532242 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 3-like [Jatropha curcas] |