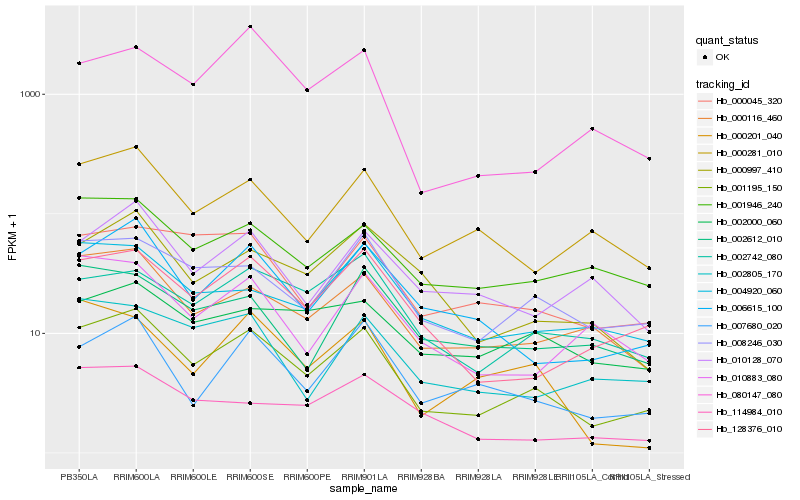
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001195_150 |
0.0 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 2 |
Hb_006615_100 |
0.1237219365 |
- |
- |
PREDICTED: uncharacterized protein LOC105640683 isoform X1 [Jatropha curcas] |
| 3 |
Hb_128376_010 |
0.1339706749 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000116_460 |
0.1340362908 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 4 [Jatropha curcas] |
| 5 |
Hb_008246_030 |
0.1353965858 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 6 |
Hb_000281_010 |
0.1431314376 |
- |
- |
hypothetical protein JCGZ_16194 [Jatropha curcas] |
| 7 |
Hb_007680_020 |
0.1479218845 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000045_320 |
0.1525070566 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000997_410 |
0.1527616177 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 10 |
Hb_001946_240 |
0.1550617786 |
- |
- |
PREDICTED: chaperone protein dnaJ 49-like [Jatropha curcas] |
| 11 |
Hb_002742_080 |
0.1569903457 |
- |
- |
unknown [Populus trichocarpa] |
| 12 |
Hb_004920_060 |
0.1614471552 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 13 |
Hb_002805_170 |
0.1617725839 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 14 |
Hb_080147_080 |
0.1651669217 |
- |
- |
peroxiredoxin, putative [Ricinus communis] |
| 15 |
Hb_010883_080 |
0.1656862733 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHR1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010128_070 |
0.1665920912 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_114984_010 |
0.1667541992 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002000_060 |
0.1721098564 |
- |
- |
PREDICTED: protein LIKE COV 2 [Jatropha curcas] |
| 19 |
Hb_000201_040 |
0.172155335 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 20 |
Hb_002612_010 |
0.1733090125 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |