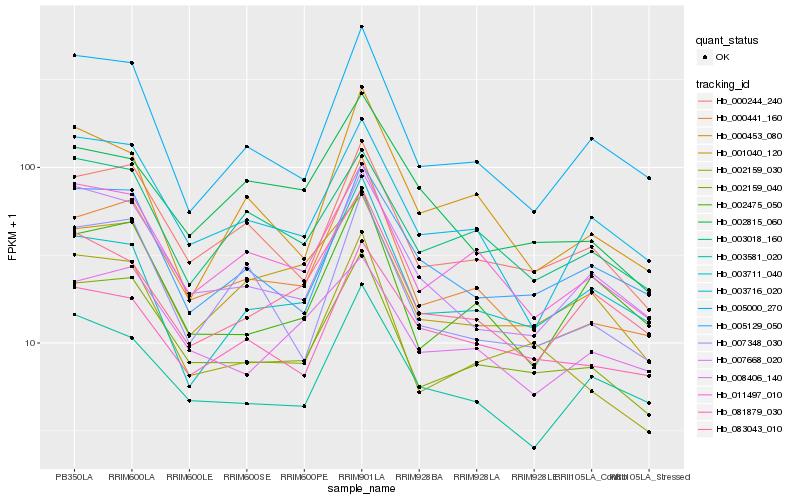
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000441_160 |
0.0 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 2 |
Hb_002159_040 |
0.0884656761 |
- |
- |
hypothetical protein PRUPE_ppa001563mg [Prunus persica] |
| 3 |
Hb_011497_010 |
0.1022090889 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 4 |
Hb_000244_240 |
0.1058542094 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_005000_270 |
0.1105160023 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_003716_020 |
0.1114628561 |
- |
- |
PREDICTED: putative vesicle-associated membrane protein 726 [Jatropha curcas] |
| 7 |
Hb_007348_030 |
0.1166538914 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002475_050 |
0.119716321 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 9 |
Hb_001040_120 |
0.1237942276 |
- |
- |
allantoinase, putative [Ricinus communis] |
| 10 |
Hb_000453_080 |
0.124100635 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_003581_020 |
0.1270501103 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007668_020 |
0.1270512175 |
- |
- |
PREDICTED: uncharacterized protein LOC105631102 [Jatropha curcas] |
| 13 |
Hb_002159_030 |
0.1285454929 |
- |
- |
AP-1 complex subunit gamma-2, putative [Ricinus communis] |
| 14 |
Hb_003711_040 |
0.1297217095 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 15 |
Hb_008406_140 |
0.1302559245 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 16 |
Hb_081879_030 |
0.1309892554 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 21 homolog [Jatropha curcas] |
| 17 |
Hb_083043_010 |
0.1310993317 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 18 |
Hb_002815_060 |
0.131387709 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 19 |
Hb_005129_050 |
0.1321357401 |
- |
- |
PREDICTED: probable 28S rRNA (cytosine(4447)-C(5))-methyltransferase [Jatropha curcas] |
| 20 |
Hb_003018_160 |
0.1352191761 |
- |
- |
PREDICTED: uncharacterized protein LOC105628876 [Jatropha curcas] |