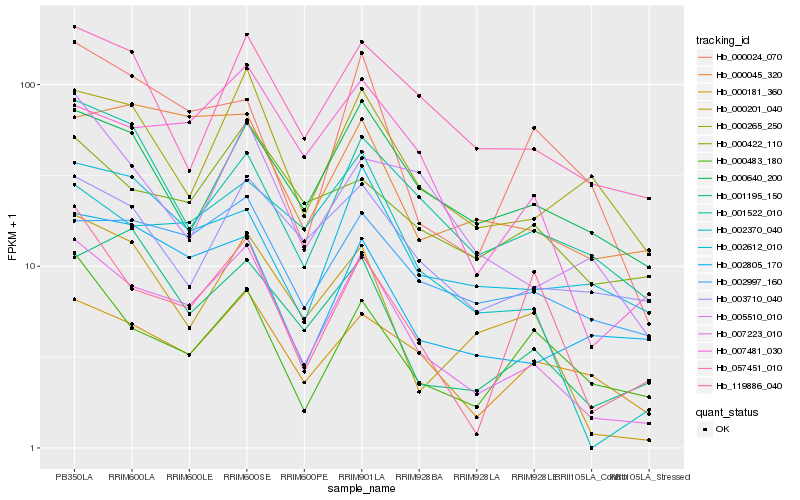
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007223_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007481_030 |
0.1420101489 |
- |
- |
PREDICTED: uncharacterized protein LOC105628433 [Jatropha curcas] |
| 3 |
Hb_002370_040 |
0.160948293 |
- |
- |
PREDICTED: uncharacterized protein LOC105644336 [Jatropha curcas] |
| 4 |
Hb_000201_040 |
0.162808102 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 5 |
Hb_000181_360 |
0.164523355 |
- |
- |
PREDICTED: uncharacterized protein LOC105644273 [Jatropha curcas] |
| 6 |
Hb_002805_170 |
0.1693210049 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 7 |
Hb_000265_250 |
0.1715848981 |
- |
- |
auxilin, putative [Ricinus communis] |
| 8 |
Hb_002997_160 |
0.1727444577 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X2 [Jatropha curcas] |
| 9 |
Hb_057451_010 |
0.1780116907 |
- |
- |
PREDICTED: uncharacterized protein LOC105635588 [Jatropha curcas] |
| 10 |
Hb_000024_070 |
0.1786142389 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000483_180 |
0.1795199342 |
- |
- |
- |
| 12 |
Hb_119886_040 |
0.1805106125 |
- |
- |
- |
| 13 |
Hb_001522_010 |
0.1811021016 |
- |
- |
PREDICTED: nucleolar protein 14 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002612_010 |
0.1812878631 |
- |
- |
hypothetical protein JCGZ_19269 [Jatropha curcas] |
| 15 |
Hb_001195_150 |
0.1816404653 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 16 |
Hb_000045_320 |
0.1825238247 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005510_010 |
0.1828734929 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000640_200 |
0.184490968 |
- |
- |
hypothetical protein RCOM_1346600 [Ricinus communis] |
| 19 |
Hb_003710_040 |
0.1856149478 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 20 |
Hb_000422_110 |
0.1856202473 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |