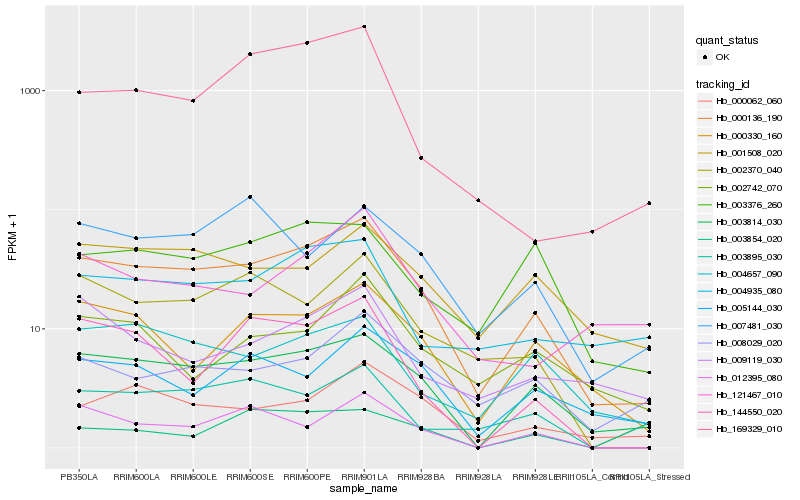
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000136_190 |
0.0 |
- |
- |
PREDICTED: protein-S-isoprenylcysteine O-methyltransferase A-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_003814_030 |
0.1035010848 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002370_040 |
0.1538070952 |
- |
- |
PREDICTED: uncharacterized protein LOC105644336 [Jatropha curcas] |
| 4 |
Hb_000330_160 |
0.1556351139 |
- |
- |
hypothetical protein JCGZ_04462 [Jatropha curcas] |
| 5 |
Hb_000062_060 |
0.1568788549 |
transcription factor |
TF Family: Orphans |
multi-sensor hybrid histidine kinase [Plautia stali symbiont] |
| 6 |
Hb_001508_020 |
0.1703778661 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 7 |
Hb_004657_090 |
0.1723665463 |
- |
- |
Chromosome-associated kinesin KLP1, putative [Ricinus communis] |
| 8 |
Hb_004935_080 |
0.1737609826 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 9 |
Hb_008029_020 |
0.1743943365 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 10 |
Hb_005144_030 |
0.1749375357 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 11 |
Hb_012395_080 |
0.1793276848 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19890 [Jatropha curcas] |
| 12 |
Hb_003376_260 |
0.1796805415 |
- |
- |
PREDICTED: uncharacterized protein LOC105642207 isoform X1 [Jatropha curcas] |
| 13 |
Hb_144550_020 |
0.1828120963 |
- |
- |
- |
| 14 |
Hb_009119_030 |
0.1877947841 |
- |
- |
GMFP4 [Medicago truncatula] |
| 15 |
Hb_003895_030 |
0.1883279043 |
- |
- |
- |
| 16 |
Hb_002742_070 |
0.1889981931 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 17 |
Hb_003854_020 |
0.1897245352 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_121467_010 |
0.1897339412 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 19 |
Hb_169329_010 |
0.1899180672 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 20 |
Hb_007481_030 |
0.1919862707 |
- |
- |
PREDICTED: uncharacterized protein LOC105628433 [Jatropha curcas] |