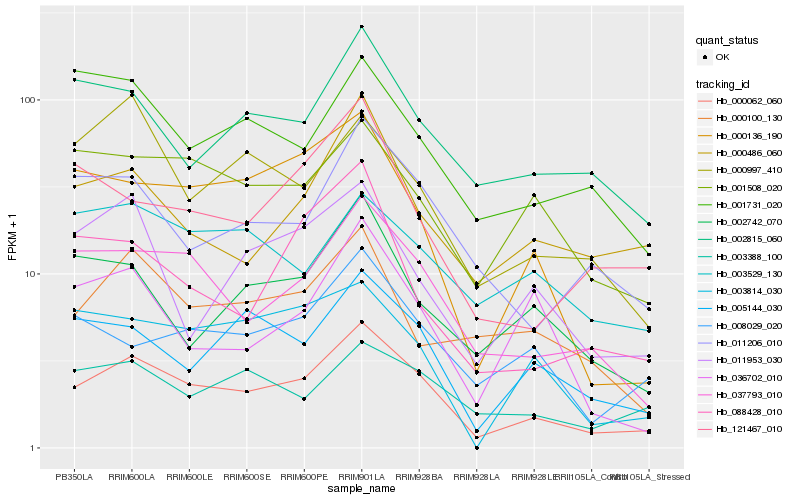
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_060 |
0.0 |
transcription factor |
TF Family: Orphans |
multi-sensor hybrid histidine kinase [Plautia stali symbiont] |
| 2 |
Hb_037793_010 |
0.1292529533 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like [Jatropha curcas] |
| 3 |
Hb_005144_030 |
0.1554433016 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 4 |
Hb_000136_190 |
0.1568788549 |
- |
- |
PREDICTED: protein-S-isoprenylcysteine O-methyltransferase A-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_011206_010 |
0.1607570419 |
- |
- |
PREDICTED: something about silencing protein 10 isoform X2 [Vitis vinifera] |
| 6 |
Hb_001508_020 |
0.1608942973 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 7 |
Hb_002815_060 |
0.1634079636 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 8 |
Hb_008029_020 |
0.1654875902 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 9 |
Hb_000486_060 |
0.1711279587 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like isoform X2 [Glycine max] |
| 10 |
Hb_003388_100 |
0.1719352044 |
- |
- |
16S rRNA processing protein RimM [Psychrobacter phenylpyruvicus] |
| 11 |
Hb_003814_030 |
0.1719792472 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_002742_070 |
0.17323081 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 13 |
Hb_000100_130 |
0.1734126062 |
- |
- |
hypothetical protein VITISV_026702 [Vitis vinifera] |
| 14 |
Hb_011953_030 |
0.174316276 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0003s14450g [Populus trichocarpa] |
| 15 |
Hb_121467_010 |
0.1773211919 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 16 |
Hb_000997_410 |
0.1779939441 |
- |
- |
PREDICTED: uncharacterized protein LOC105632066 [Jatropha curcas] |
| 17 |
Hb_001731_020 |
0.1802015157 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 18 |
Hb_003529_130 |
0.1826113488 |
- |
- |
PREDICTED: ribosome biogenesis protein BOP1 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_036702_010 |
0.1840266182 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 20 |
Hb_088428_010 |
0.1862325849 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |