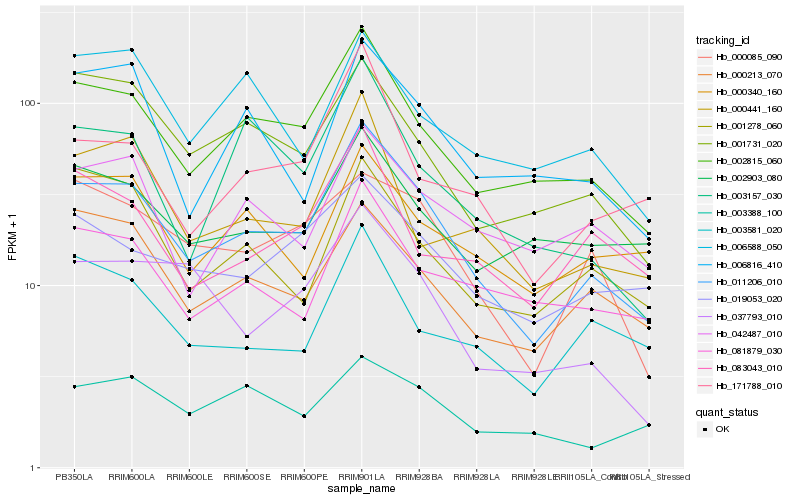
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011206_010 |
0.0 |
- |
- |
PREDICTED: something about silencing protein 10 isoform X2 [Vitis vinifera] |
| 2 |
Hb_002815_060 |
0.0958930461 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 3 |
Hb_001731_020 |
0.1357046029 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 4 |
Hb_000213_070 |
0.136799019 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000085_090 |
0.1386138415 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 6 |
Hb_171788_010 |
0.1396689413 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 7 |
Hb_019053_020 |
0.1426265687 |
- |
- |
hypothetical protein MIMGU_mgv1a0105441mg, partial [Erythranthe guttata] |
| 8 |
Hb_081879_030 |
0.144550493 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 21 homolog [Jatropha curcas] |
| 9 |
Hb_042487_010 |
0.1456449936 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 9-like [Jatropha curcas] |
| 10 |
Hb_000441_160 |
0.1458124807 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 11 |
Hb_037793_010 |
0.146102268 |
- |
- |
PREDICTED: microtubule-associated protein 70-2-like [Jatropha curcas] |
| 12 |
Hb_006816_410 |
0.1479379267 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit A [Jatropha curcas] |
| 13 |
Hb_003157_030 |
0.1489583089 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Nicotiana sylvestris] |
| 14 |
Hb_001278_060 |
0.149050874 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 15 |
Hb_000340_160 |
0.1497507543 |
- |
- |
PREDICTED: elongation factor Tu, mitochondrial [Jatropha curcas] |
| 16 |
Hb_002903_080 |
0.1498923179 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 17 |
Hb_083043_010 |
0.1503527014 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 18 |
Hb_003581_020 |
0.1520591077 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003388_100 |
0.1540706856 |
- |
- |
16S rRNA processing protein RimM [Psychrobacter phenylpyruvicus] |
| 20 |
Hb_006588_050 |
0.1564308264 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |