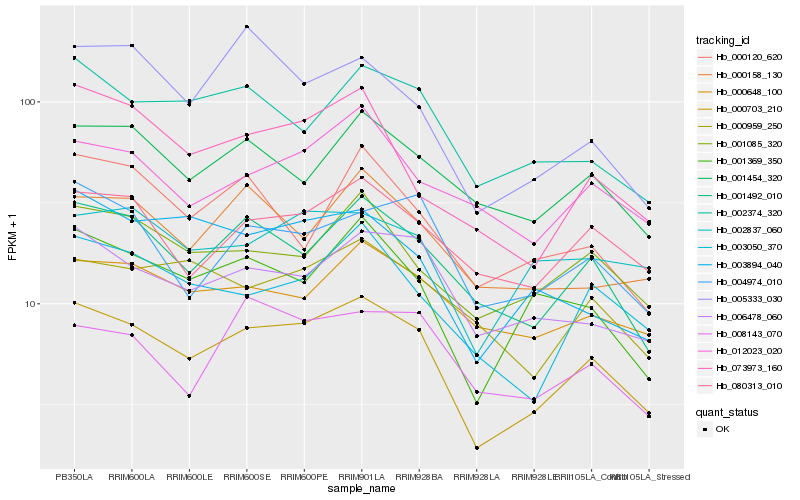
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_210 |
0.0 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 2 |
Hb_001492_010 |
0.0971756226 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like isoform X2 [Populus euphratica] |
| 3 |
Hb_001369_350 |
0.104821132 |
- |
- |
Cisplatin resistance-associated overexpressed protein, putative [Ricinus communis] |
| 4 |
Hb_003050_370 |
0.1114673874 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 5 |
Hb_006478_060 |
0.1115601525 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001085_320 |
0.1155513531 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_002374_320 |
0.1158639058 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 8 |
Hb_080313_010 |
0.1159374587 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 9 |
Hb_004974_010 |
0.116616078 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Prunus mume] |
| 10 |
Hb_073973_160 |
0.1169738789 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 11 |
Hb_003894_040 |
0.1230192449 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 12 |
Hb_001454_320 |
0.123246004 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002837_060 |
0.1244898417 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 14 |
Hb_005333_030 |
0.1260817583 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 15 |
Hb_000959_250 |
0.1262518168 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase large subunit [Jatropha curcas] |
| 16 |
Hb_000158_130 |
0.126434518 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000648_100 |
0.1265993352 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 18 |
Hb_012023_020 |
0.1273993704 |
- |
- |
hypothetical protein JCGZ_03076 [Jatropha curcas] |
| 19 |
Hb_000120_620 |
0.1301941341 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 20 |
Hb_008143_070 |
0.1309697296 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |