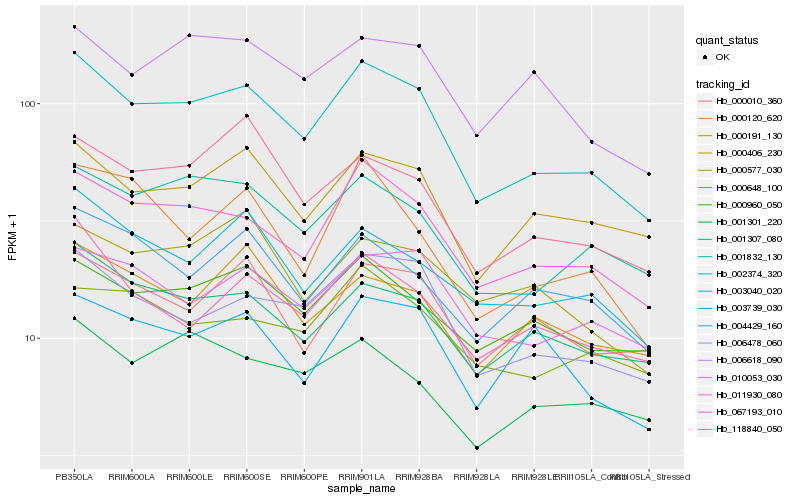
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_320 |
0.0 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 2 |
Hb_011930_080 |
0.0555378609 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 3 |
Hb_006478_060 |
0.0657957025 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001832_130 |
0.0780148069 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 5 |
Hb_001307_080 |
0.0796883027 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 6 |
Hb_000406_230 |
0.0812058575 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 7 |
Hb_003040_020 |
0.0834490898 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 8 |
Hb_010053_030 |
0.0835101747 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 9 |
Hb_000960_050 |
0.0835370227 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 10 |
Hb_118840_050 |
0.0847160013 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |
| 11 |
Hb_067193_010 |
0.0870646643 |
- |
- |
- |
| 12 |
Hb_000010_360 |
0.0872955965 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 13 |
Hb_004429_160 |
0.0876214632 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_006618_090 |
0.0902368076 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 15 |
Hb_000120_620 |
0.090952739 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 16 |
Hb_000648_100 |
0.0916113412 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 17 |
Hb_000191_130 |
0.0926235256 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 18 |
Hb_003739_030 |
0.0938944986 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 19 |
Hb_000577_030 |
0.0942752129 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase 12 -like protein [Gossypium arboreum] |
| 20 |
Hb_001301_220 |
0.0946770598 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |