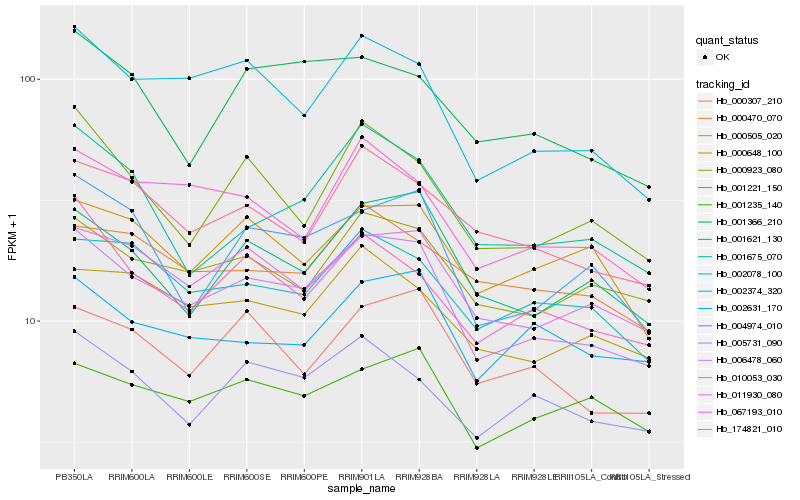
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006478_060 |
0.0 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002374_320 |
0.0657957025 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 3 |
Hb_010053_030 |
0.0684686718 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 4 |
Hb_011930_080 |
0.0800053267 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 5 |
Hb_000923_080 |
0.0812111751 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 6 |
Hb_001621_130 |
0.0816190423 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_174821_010 |
0.0827928387 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 14 [Jatropha curcas] |
| 8 |
Hb_002078_100 |
0.0834042981 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 9 |
Hb_000307_210 |
0.0845275618 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 10 |
Hb_000648_100 |
0.0847410521 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 11 |
Hb_004974_010 |
0.0861429431 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Prunus mume] |
| 12 |
Hb_067193_010 |
0.0873270775 |
- |
- |
- |
| 13 |
Hb_005731_090 |
0.0883025202 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 14 |
Hb_001221_150 |
0.0886494057 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 15 |
Hb_000505_020 |
0.0925909412 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 16 |
Hb_001235_140 |
0.092694473 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 17 |
Hb_001675_070 |
0.0943472993 |
- |
- |
eukaryotic translation elongation factor, putative [Ricinus communis] |
| 18 |
Hb_002631_170 |
0.0943883426 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 19 |
Hb_001366_210 |
0.0944868022 |
- |
- |
cathepsin B, putative [Ricinus communis] |
| 20 |
Hb_000470_070 |
0.0949103951 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |