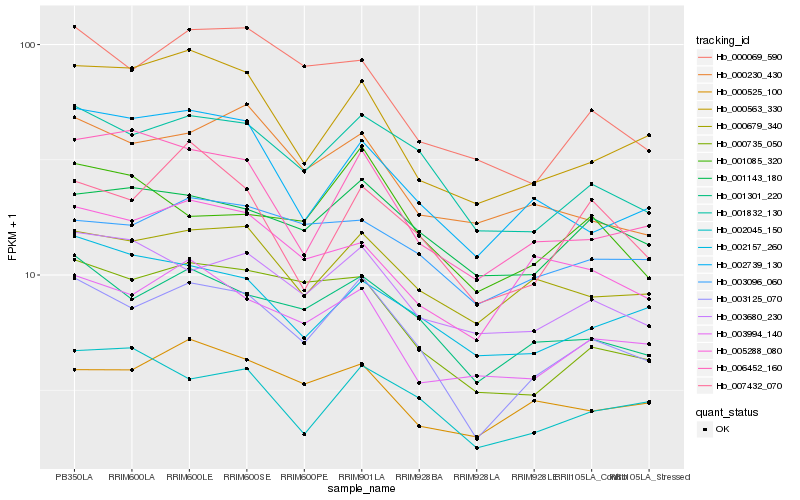
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003125_070 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio1-like [Jatropha curcas] |
| 2 |
Hb_001832_130 |
0.0800044409 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 3 |
Hb_001301_220 |
0.0804528795 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 4 |
Hb_000735_050 |
0.1015923693 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 5 |
Hb_003994_140 |
0.1017891011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000563_330 |
0.1018707435 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_000679_340 |
0.1035870309 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003680_230 |
0.1066316256 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 9 |
Hb_006452_160 |
0.1080517156 |
- |
- |
structural constituent of ribosome, putative [Ricinus communis] |
| 10 |
Hb_002739_130 |
0.1087789913 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Populus euphratica] |
| 11 |
Hb_000069_590 |
0.110539572 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 12 |
Hb_007432_070 |
0.110576912 |
- |
- |
PREDICTED: glutamine--tRNA ligase [Jatropha curcas] |
| 13 |
Hb_001143_180 |
0.1110488059 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 14 |
Hb_005288_080 |
0.1114056653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000525_100 |
0.1120141348 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 16 |
Hb_002045_150 |
0.1128797299 |
- |
- |
histone acetyltransferase gcn5, putative [Ricinus communis] |
| 17 |
Hb_002157_260 |
0.1155245836 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 18 |
Hb_003096_060 |
0.1158146853 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 19 |
Hb_000230_430 |
0.1162703981 |
- |
- |
catalytic, putative [Ricinus communis] |
| 20 |
Hb_001085_320 |
0.1170032734 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |