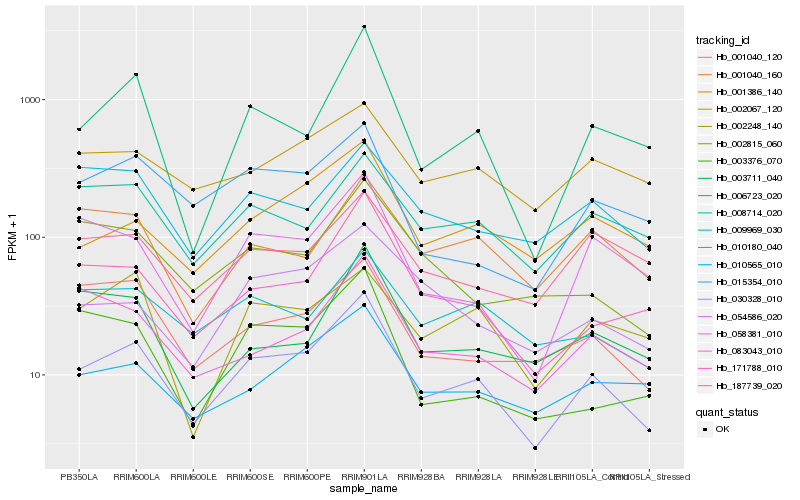
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030328_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10015936mg [Citrus clementina] |
| 2 |
Hb_171788_010 |
0.1315466165 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 3 |
Hb_006723_020 |
0.1349539911 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 4 |
Hb_001386_140 |
0.1379193955 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 5 |
Hb_010565_010 |
0.1386169122 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 6 |
Hb_058381_010 |
0.1398233659 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 7 |
Hb_054586_020 |
0.1456386578 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 8 |
Hb_008714_020 |
0.1478476629 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 9 |
Hb_001040_120 |
0.1482758627 |
- |
- |
allantoinase, putative [Ricinus communis] |
| 10 |
Hb_015354_010 |
0.1487587148 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 11 |
Hb_002067_120 |
0.1509613396 |
- |
- |
PREDICTED: uncharacterized protein LOC105111554 isoform X1 [Populus euphratica] |
| 12 |
Hb_083043_010 |
0.1515397007 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 13 |
Hb_009969_030 |
0.1519565484 |
- |
- |
PREDICTED: argininosuccinate synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001040_160 |
0.1525369022 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 15 |
Hb_010180_040 |
0.1553141638 |
- |
- |
PREDICTED: prosaposin isoform X1 [Jatropha curcas] |
| 16 |
Hb_187739_020 |
0.1577990711 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 17 |
Hb_002815_060 |
0.1589537173 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 18 |
Hb_003711_040 |
0.160959398 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 19 |
Hb_003376_070 |
0.1620526748 |
- |
- |
hypothetical protein B456_009G419800 [Gossypium raimondii] |
| 20 |
Hb_002248_140 |
0.1631549542 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |