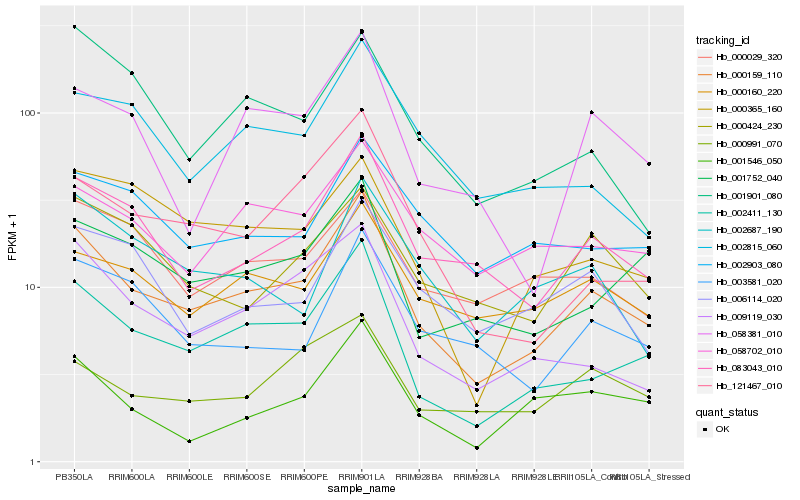
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000159_110 |
0.0 |
- |
- |
PREDICTED: thioredoxin-like protein YLS8 isoform X2 [Malus domestica] |
| 2 |
Hb_002411_130 |
0.0973536399 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Populus euphratica] |
| 3 |
Hb_083043_010 |
0.117336554 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 4 |
Hb_009119_030 |
0.1342454033 |
- |
- |
GMFP4 [Medicago truncatula] |
| 5 |
Hb_000160_220 |
0.1384941739 |
- |
- |
PREDICTED: protein AAR2 homolog isoform X1 [Jatropha curcas] |
| 6 |
Hb_001752_040 |
0.1406699534 |
- |
- |
hypothetical protein EUGRSUZ_C02580 [Eucalyptus grandis] |
| 7 |
Hb_058702_010 |
0.1410103434 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_121467_010 |
0.1418069971 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 9 |
Hb_058381_010 |
0.1445873439 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 10 |
Hb_003581_020 |
0.1457207953 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002903_080 |
0.1484647089 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 12 |
Hb_001546_050 |
0.1489847057 |
- |
- |
- |
| 13 |
Hb_000365_160 |
0.149001314 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 14 |
Hb_000991_070 |
0.149119911 |
- |
- |
phytochrome A specific signal transduction component family protein [Populus trichocarpa] |
| 15 |
Hb_002815_060 |
0.1491208412 |
- |
- |
hypothetical protein B456_002G110500 [Gossypium raimondii] |
| 16 |
Hb_000029_320 |
0.1491249122 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 17 |
Hb_000424_230 |
0.150126999 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Glycine max] |
| 18 |
Hb_006114_020 |
0.1526575265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002687_190 |
0.1530058219 |
- |
- |
prolyl oligopeptidase [Cicer arietinum] |
| 20 |
Hb_001901_080 |
0.1539296976 |
- |
- |
PREDICTED: xylose isomerase [Jatropha curcas] |