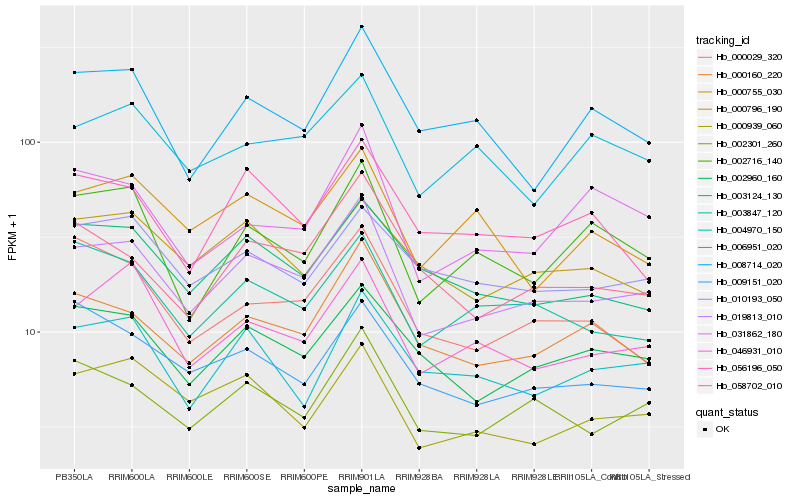
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019813_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646947 [Jatropha curcas] |
| 2 |
Hb_002301_260 |
0.0780807283 |
- |
- |
hypothetical protein MIMGU_mgv1a0031591mg, partial [Erythranthe guttata] |
| 3 |
Hb_000160_220 |
0.088368956 |
- |
- |
PREDICTED: protein AAR2 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_002960_160 |
0.091847111 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 5 |
Hb_046931_010 |
0.0942737986 |
- |
- |
hypothetical protein JCGZ_26924 [Jatropha curcas] |
| 6 |
Hb_000939_060 |
0.094846986 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 7 |
Hb_003847_120 |
0.0953841622 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_003124_130 |
0.0993933491 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 9 |
Hb_058702_010 |
0.1001699559 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_006951_020 |
0.1039053439 |
- |
- |
Rop3 [Hevea brasiliensis] |
| 11 |
Hb_002716_140 |
0.1042727592 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 12 |
Hb_031862_180 |
0.1064274096 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 13 |
Hb_010193_050 |
0.1073761041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009151_020 |
0.108637333 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 15 |
Hb_008714_020 |
0.1091482003 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 16 |
Hb_004970_150 |
0.1092170013 |
- |
- |
PREDICTED: uncharacterized protein LOC105633636 [Jatropha curcas] |
| 17 |
Hb_000796_190 |
0.1094404752 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 18 |
Hb_056196_050 |
0.1095965171 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 19 |
Hb_000755_030 |
0.1097815157 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 20 |
Hb_000029_320 |
0.1102929661 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |