| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
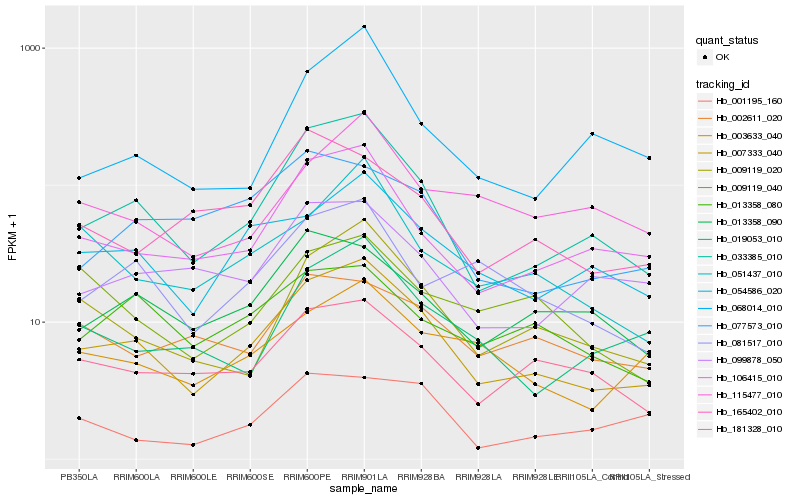
Hb_007333_040 |
0.0 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 2 |
Hb_033385_010 |
0.0863804952 |
- |
- |
hypothetical protein JCGZ_06094 [Jatropha curcas] |
| 3 |
Hb_106415_010 |
0.1303286237 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 4 |
Hb_009119_020 |
0.1331629974 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 5 |
Hb_054586_020 |
0.1447907527 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 6 |
Hb_181328_010 |
0.1516173374 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_019053_010 |
0.1585889303 |
- |
- |
- |
| 8 |
Hb_003633_040 |
0.1625361597 |
- |
- |
hypothetical protein B456_008G146200 [Gossypium raimondii] |
| 9 |
Hb_051437_010 |
0.1650950471 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 10 |
Hb_081517_010 |
0.1664118571 |
- |
- |
PREDICTED: putative potassium transporter 12 isoform X2 [Jatropha curcas] |
| 11 |
Hb_013358_090 |
0.166563444 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 12 |
Hb_099878_050 |
0.1716213679 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 13 |
Hb_068014_010 |
0.1725239416 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 14 |
Hb_013358_080 |
0.1764911853 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 15 |
Hb_115477_010 |
0.1776853077 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 16 |
Hb_077573_010 |
0.1839478121 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_009119_040 |
0.1840778737 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2A [Jatropha curcas] |
| 18 |
Hb_001195_160 |
0.1845884013 |
- |
- |
- |
| 19 |
Hb_002611_020 |
0.1846398176 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 20 |
Hb_165402_010 |
0.1875713292 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |