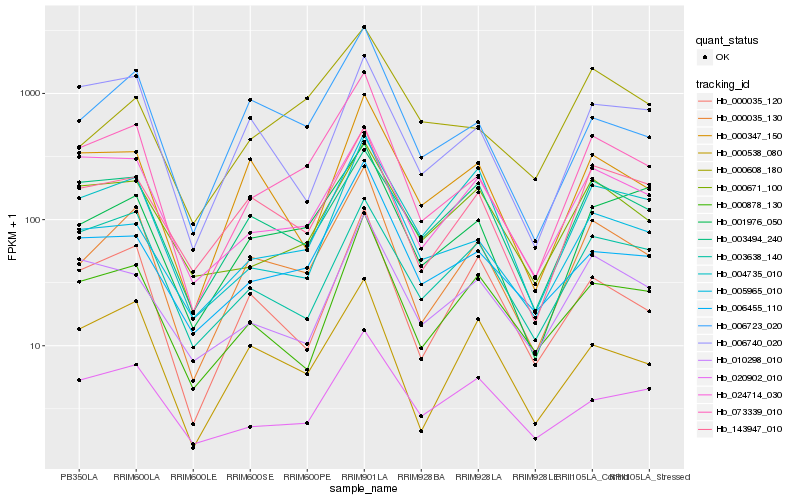
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_130 |
0.0 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 2 |
Hb_073339_010 |
0.1065863712 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 3 |
Hb_006723_020 |
0.1179980649 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 4 |
Hb_143947_010 |
0.1226783655 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X2 [Pyrus x bretschneideri] |
| 5 |
Hb_000035_120 |
0.1274503817 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 6 |
Hb_000538_080 |
0.1349092201 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 7 |
Hb_000878_130 |
0.1368361436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001976_050 |
0.1376588131 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 9 |
Hb_000347_150 |
0.1485015101 |
- |
- |
Nucleolysin TIAR [Morus notabilis] |
| 10 |
Hb_005965_010 |
0.15674829 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 11 |
Hb_006455_110 |
0.1583793043 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_003494_240 |
0.1624831788 |
- |
- |
PREDICTED: uncharacterized protein LOC105644075 [Jatropha curcas] |
| 13 |
Hb_024714_030 |
0.1640644277 |
- |
- |
PREDICTED: sm-like protein LSM4 [Jatropha curcas] |
| 14 |
Hb_003638_140 |
0.167932864 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_020902_010 |
0.1682146719 |
- |
- |
PREDICTED: uncharacterized protein LOC104595551 [Nelumbo nucifera] |
| 16 |
Hb_000671_100 |
0.1708951041 |
- |
- |
hypothetical protein POPTR_0017s06410g [Populus trichocarpa] |
| 17 |
Hb_010298_010 |
0.1713793344 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 [Jatropha curcas] |
| 18 |
Hb_004735_010 |
0.1726192007 |
- |
- |
PREDICTED: protein arginine N-methyltransferase PRMT10 [Vitis vinifera] |
| 19 |
Hb_006740_020 |
0.1727943895 |
- |
- |
PREDICTED: bis(5'-adenosyl)-triphosphatase [Jatropha curcas] |
| 20 |
Hb_000608_180 |
0.1741080314 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Tarenaya hassleriana] |