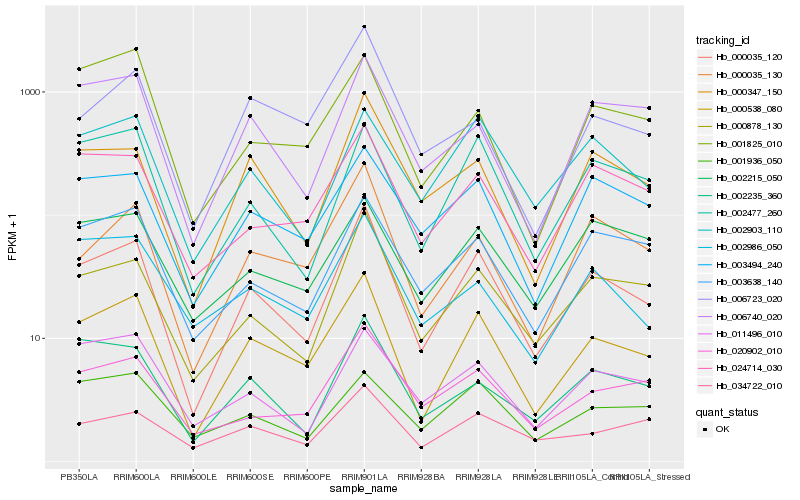
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000538_080 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_000035_120 |
0.077839995 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 3 |
Hb_000035_130 |
0.1349092201 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 4 |
Hb_003494_240 |
0.1465944879 |
- |
- |
PREDICTED: uncharacterized protein LOC105644075 [Jatropha curcas] |
| 5 |
Hb_024714_030 |
0.1467620554 |
- |
- |
PREDICTED: sm-like protein LSM4 [Jatropha curcas] |
| 6 |
Hb_002477_260 |
0.1480407926 |
- |
- |
PREDICTED: protein phosphatase 2C 16 [Jatropha curcas] |
| 7 |
Hb_000878_130 |
0.1487558669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003638_140 |
0.1511386715 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_001825_010 |
0.151206635 |
- |
- |
PREDICTED: bet1-like SNARE 1-1 [Jatropha curcas] |
| 10 |
Hb_006740_020 |
0.1538513034 |
- |
- |
PREDICTED: bis(5'-adenosyl)-triphosphatase [Jatropha curcas] |
| 11 |
Hb_000347_150 |
0.1568492002 |
- |
- |
Nucleolysin TIAR [Morus notabilis] |
| 12 |
Hb_006723_020 |
0.1576114852 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 13 |
Hb_020902_010 |
0.1590847075 |
- |
- |
PREDICTED: uncharacterized protein LOC104595551 [Nelumbo nucifera] |
| 14 |
Hb_002235_360 |
0.1613294358 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 15 |
Hb_002986_050 |
0.1621874989 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 16 |
Hb_002903_110 |
0.1623843185 |
- |
- |
PREDICTED: RNA polymerase-associated protein RTF1 homolog [Jatropha curcas] |
| 17 |
Hb_002215_050 |
0.1626044804 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 18 |
Hb_011496_010 |
0.1639688451 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g38420, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001936_050 |
0.1644547468 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210-like [Prunus mume] |
| 20 |
Hb_034722_010 |
0.1646090299 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g13770, mitochondrial [Populus euphratica] |