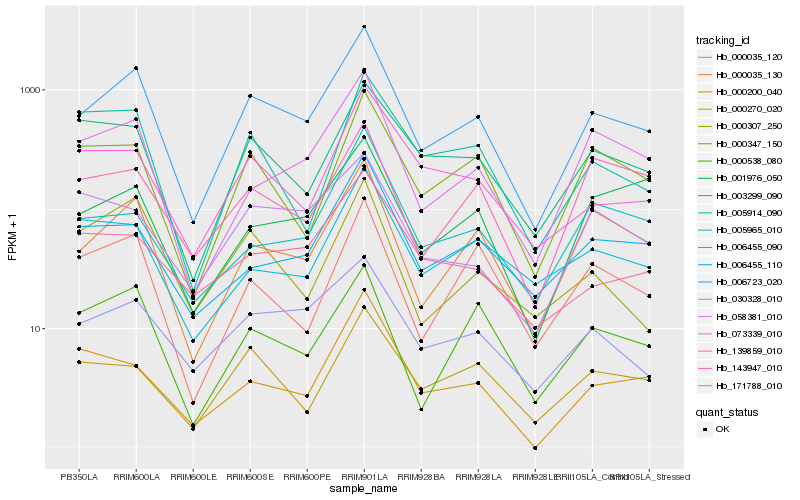
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006723_020 |
0.0 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 2 |
Hb_000035_130 |
0.1179980649 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 3 |
Hb_030328_010 |
0.1349539911 |
- |
- |
hypothetical protein CICLE_v10015936mg [Citrus clementina] |
| 4 |
Hb_073339_010 |
0.1390097512 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 5 |
Hb_006455_110 |
0.1502101483 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000347_150 |
0.1508778504 |
- |
- |
Nucleolysin TIAR [Morus notabilis] |
| 7 |
Hb_139859_010 |
0.1537173549 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 8 |
Hb_003299_090 |
0.1552682319 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |
| 9 |
Hb_000538_080 |
0.1576114852 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 10 |
Hb_000270_020 |
0.1598200802 |
- |
- |
hypothetical protein POPTR_0015s07630g [Populus trichocarpa] |
| 11 |
Hb_171788_010 |
0.1605689485 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 12 |
Hb_000035_120 |
0.1612227281 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 13 |
Hb_005965_010 |
0.1617189735 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 14 |
Hb_006455_090 |
0.1651992131 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 15 |
Hb_001976_050 |
0.1671439989 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 16 |
Hb_000307_250 |
0.1688908693 |
- |
- |
PREDICTED: probable nucleoredoxin 2 [Jatropha curcas] |
| 17 |
Hb_005914_090 |
0.1730877981 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 18 |
Hb_058381_010 |
0.1735524877 |
- |
- |
hypothetical protein JCGZ_13280 [Jatropha curcas] |
| 19 |
Hb_143947_010 |
0.1751852223 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X2 [Pyrus x bretschneideri] |
| 20 |
Hb_000200_040 |
0.1762016386 |
- |
- |
- |