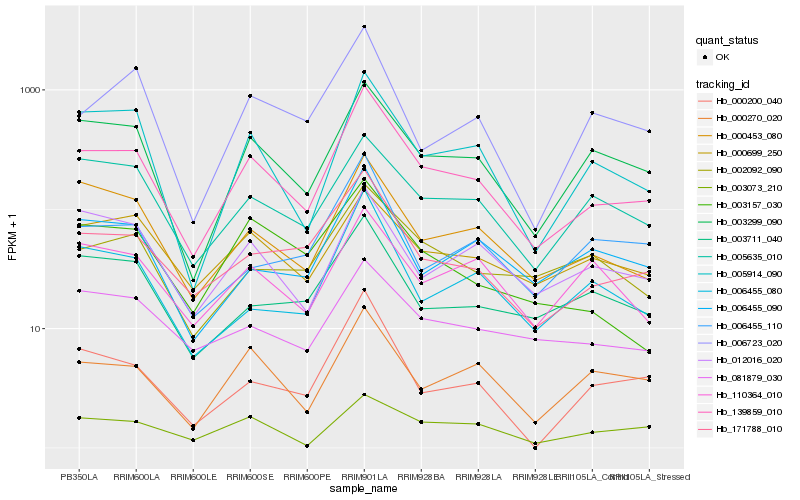
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_139859_010 |
0.0 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 2 |
Hb_171788_010 |
0.1140819716 |
- |
- |
hypothetical protein CISIN_1g007892mg [Citrus sinensis] |
| 3 |
Hb_003299_090 |
0.1148273383 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |
| 4 |
Hb_005914_090 |
0.1241769708 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 5 |
Hb_000453_080 |
0.1266799978 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000270_020 |
0.1356646985 |
- |
- |
hypothetical protein POPTR_0015s07630g [Populus trichocarpa] |
| 7 |
Hb_006455_080 |
0.1411727788 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 8 |
Hb_006455_090 |
0.142376611 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 9 |
Hb_005635_010 |
0.1478945524 |
- |
- |
PREDICTED: cleft lip and palate transmembrane protein 1 homolog [Jatropha curcas] |
| 10 |
Hb_000200_040 |
0.1513501375 |
- |
- |
- |
| 11 |
Hb_003711_040 |
0.1516981836 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 12 |
Hb_012016_020 |
0.1526151755 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105636265 [Jatropha curcas] |
| 13 |
Hb_002092_090 |
0.1535122106 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 14 |
Hb_006723_020 |
0.1537173549 |
- |
- |
ARF2-B: ADP-ribosylation factor 2-B [Gossypium arboreum] |
| 15 |
Hb_006455_110 |
0.1539669661 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_000699_250 |
0.1542253778 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 17 |
Hb_003073_210 |
0.1553131928 |
- |
- |
hypothetical protein CISIN_1g036722mg, partial [Citrus sinensis] |
| 18 |
Hb_081879_030 |
0.155406439 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 21 homolog [Jatropha curcas] |
| 19 |
Hb_003157_030 |
0.1577700903 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Nicotiana sylvestris] |
| 20 |
Hb_110364_010 |
0.1589323699 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |