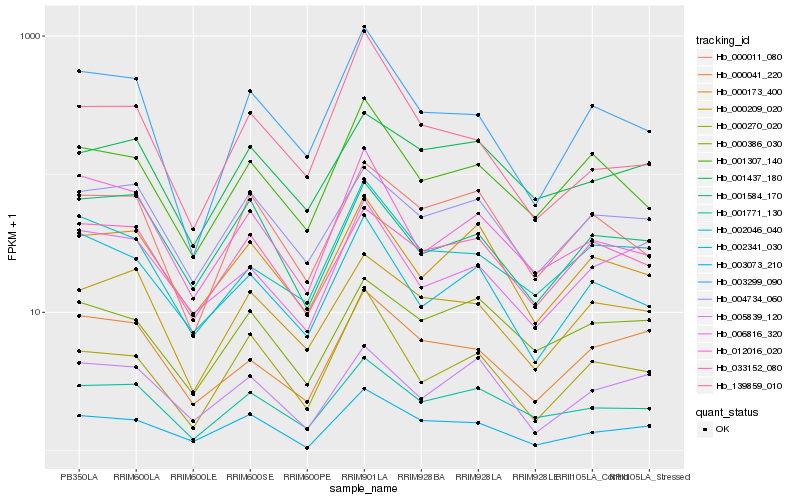
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003073_210 |
0.0 |
- |
- |
hypothetical protein CISIN_1g036722mg, partial [Citrus sinensis] |
| 2 |
Hb_000270_020 |
0.1315490143 |
- |
- |
hypothetical protein POPTR_0015s07630g [Populus trichocarpa] |
| 3 |
Hb_001771_130 |
0.1377514985 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g27610 [Jatropha curcas] |
| 4 |
Hb_000041_220 |
0.1416136375 |
- |
- |
polyribonucleotide nucleotidyltransferase, putative [Ricinus communis] |
| 5 |
Hb_003299_090 |
0.1441634725 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |
| 6 |
Hb_001584_170 |
0.1450337832 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 7 |
Hb_005839_120 |
0.1479216461 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 8 |
Hb_000173_400 |
0.148421206 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18950 [Jatropha curcas] |
| 9 |
Hb_000011_080 |
0.1488701604 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 10 |
Hb_000209_020 |
0.1490780845 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 11 |
Hb_006816_320 |
0.1494727395 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 16-like [Jatropha curcas] |
| 12 |
Hb_004734_060 |
0.1500965022 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 13 |
Hb_033152_080 |
0.1527413048 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 14 |
Hb_012016_020 |
0.1530327746 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105636265 [Jatropha curcas] |
| 15 |
Hb_002341_030 |
0.1543292991 |
- |
- |
PREDICTED: transcription factor bHLH140 [Jatropha curcas] |
| 16 |
Hb_001307_140 |
0.1546522276 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Jatropha curcas] |
| 17 |
Hb_000386_030 |
0.1552719868 |
- |
- |
PREDICTED: uncharacterized protein LOC105629270 [Jatropha curcas] |
| 18 |
Hb_139859_010 |
0.1553131928 |
- |
- |
PREDICTED: cell division control protein 48 homolog D-like [Nicotiana sylvestris] |
| 19 |
Hb_001437_180 |
0.1558417123 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 20 |
Hb_002046_040 |
0.1567458175 |
- |
- |
PREDICTED: topless-related protein 4 [Malus domestica] |