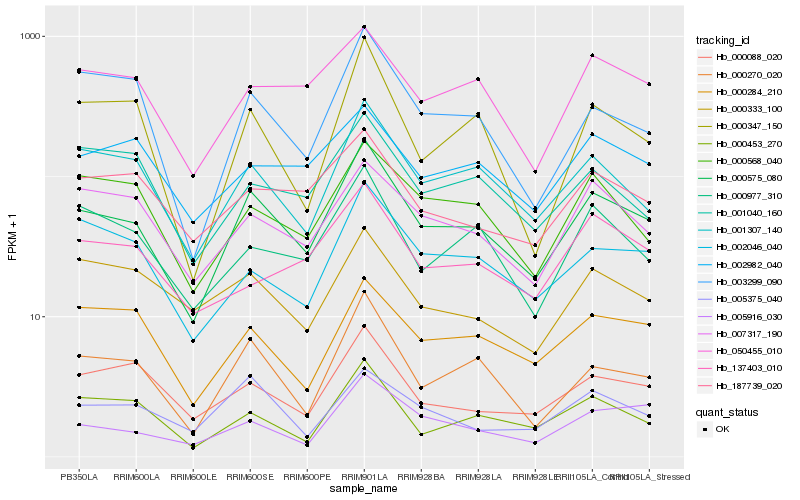
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000575_080 |
0.0 |
- |
- |
PREDICTED: CD2 antigen cytoplasmic tail-binding protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001307_140 |
0.1077566507 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Jatropha curcas] |
| 3 |
Hb_000270_020 |
0.1100121202 |
- |
- |
hypothetical protein POPTR_0015s07630g [Populus trichocarpa] |
| 4 |
Hb_050455_010 |
0.1265690657 |
- |
- |
PREDICTED: prosaposin isoform X2 [Jatropha curcas] |
| 5 |
Hb_000088_020 |
0.1331704155 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 6 |
Hb_003299_090 |
0.1335491453 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |
| 7 |
Hb_007317_190 |
0.1338646937 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 8 |
Hb_005916_030 |
0.1340548566 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 9 |
Hb_000977_310 |
0.1342810246 |
- |
- |
PREDICTED: gamma-tubulin complex component 2 isoform X4 [Jatropha curcas] |
| 10 |
Hb_002046_040 |
0.1347714177 |
- |
- |
PREDICTED: topless-related protein 4 [Malus domestica] |
| 11 |
Hb_000284_210 |
0.1349377275 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000568_040 |
0.1353942359 |
- |
- |
PREDICTED: mRNA-decapping enzyme subunit 2 [Jatropha curcas] |
| 13 |
Hb_000453_270 |
0.1384778564 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 [Propithecus coquereli] |
| 14 |
Hb_000347_150 |
0.1394458216 |
- |
- |
Nucleolysin TIAR [Morus notabilis] |
| 15 |
Hb_187739_020 |
0.1397837009 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 16 |
Hb_005375_040 |
0.1398309445 |
- |
- |
dicer-1, putative [Ricinus communis] |
| 17 |
Hb_002982_040 |
0.1441700349 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 18 |
Hb_001040_160 |
0.1478287231 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 19 |
Hb_137403_010 |
0.1485840657 |
- |
- |
PREDICTED: actin-related protein 9 [Jatropha curcas] |
| 20 |
Hb_000333_100 |
0.1486335789 |
- |
- |
PREDICTED: tubulin--tyrosine ligase-like protein 12 isoform X1 [Jatropha curcas] |