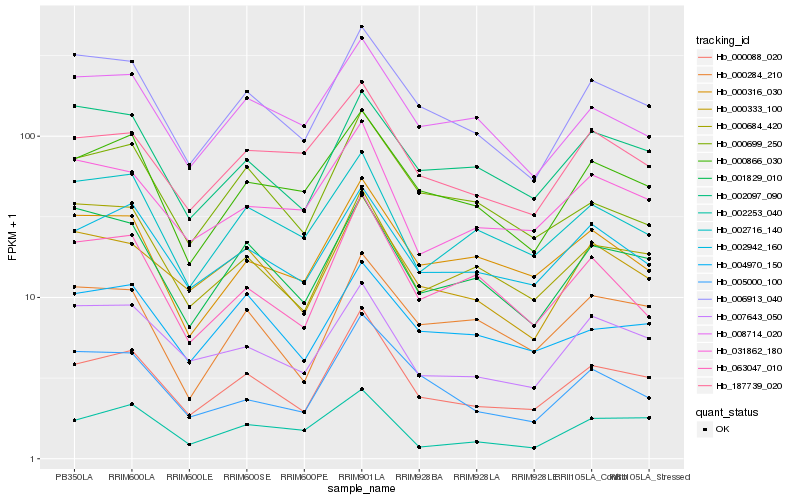
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000088_020 |
0.0 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 2 |
Hb_006913_040 |
0.0901580573 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 3 |
Hb_000333_100 |
0.0957749381 |
- |
- |
PREDICTED: tubulin--tyrosine ligase-like protein 12 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002942_160 |
0.095924123 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 5 |
Hb_000866_030 |
0.0999754343 |
- |
- |
hypothetical protein RCOM_1615820 [Ricinus communis] |
| 6 |
Hb_031862_180 |
0.1035143008 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 7 |
Hb_007643_050 |
0.1045077868 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_187739_020 |
0.1052559133 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 9 |
Hb_000316_030 |
0.1088242943 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_005000_100 |
0.1094407534 |
- |
- |
PCI domain-containing protein 2 [Morus notabilis] |
| 11 |
Hb_063047_010 |
0.1104222265 |
- |
- |
peroxisomal targeting signal type 1 (pts1) receptor, putative [Ricinus communis] |
| 12 |
Hb_001829_010 |
0.1109664255 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000284_210 |
0.1116254508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000699_250 |
0.1127851342 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 15 |
Hb_002097_090 |
0.1127897422 |
- |
- |
Splicing factor 3A subunit, putative [Ricinus communis] |
| 16 |
Hb_008714_020 |
0.113058235 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 17 |
Hb_002716_140 |
0.11330894 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 18 |
Hb_000684_420 |
0.1133145972 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_002253_040 |
0.1142828241 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 20 |
Hb_004970_150 |
0.114441753 |
- |
- |
PREDICTED: uncharacterized protein LOC105633636 [Jatropha curcas] |